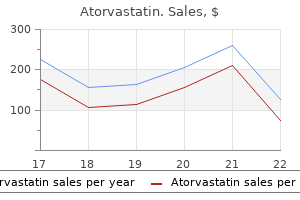
Buy discount atorvastatin 20 mg on line
They are probably homologous to the (H3-H4)2 tetramers found in the core of the eukaryotic nucleosome is there cholesterol in eggs bad for you purchase atorvastatin with mastercard. Viral genomes are smaller than most bacterial genomes and often are missing key genes for survival since they can rely on their host cell to provide these gene products. Organelle genomes are circular and only have some of the genes necessary for their function within the cell. Often, host encoded proteins are used in combination with their proteins to function effectively. A consensus sequence is determined by comparing sets of related sequences and determining which nucleotide or amino acid is most prevalent at that particular position. Shortly after replication initiation, ParA and ParB recognize the parS site near the origin and then assemble filaments that move the two chromosomes into each daughter cell. The beads or nucleosomes are then compacted into a 30 nm fiber by forming a solenoid or zigzag conformation. This form loops from a protein scaffold and then condenses even further during mitosis. The amino terminus of histones juts from the core proteins and can be modified by adding acetyl groups, methyl groups, or phosphate groups. Name the two genes that are usually found in multiple copies in a prokaryotic cell. What are the alternate structures formed due to strain caused by supercoiling and how do they differ Name the core histones and the linker histone and describe how they are arranged in a nucleosome. The researchers want to identify other genes from other flowers that might be similar in sequence. In the laboratory, a brand new single-celled eukaryotic organism was finally able to be grown in large quantities. The researchers were interested in sequencing the genome to determine if this new life form was similar to other single-celled eukaryotes. Great diversity in structure, arrangement, and numbers of chromosomes exists for all of life. Prokaryotic cells typically have a single, circular chromosome while higher organisms have multiple, linear chromosomes, each containing thousands of genes. For the purposes of this chapter, we will include viruses, even though they are considered nonliving. These complexes have to fit within the tight constraints of a prokaryotic cell or within the nucleus of a eukaryotic cell. Viral genomes are smaller than most bacterial genomes, and often are missing key genes for survival since they can rely on their host cell to provide these gene products. Operons are units of multiple coding regions that are under the control of a single transcriptional promoter. Often host encoded proteins are used in combination with their proteins to function effectively. Since many organellar genes are similar to those of bacteria, it is thought that organelles are degenerate symbiotic organisms. Some eukaryotic organelles, mainly chloroplasts and mitochondria, contain their own genomes, which are smaller and circular in nature. The genes encoding enzymes for cellular respiration (mitochondria) and photosynthesis (chloroplasts) are mostly carried on these small genomes. Upon analysis of organelle genomes, the genes present are similar to prokaryotic genes. Also, the ribosomes are more similar to bacterial ribosomes than they are to those of the eukaryotic cytoplasm in which the organelles reside. One theory to explain this, called the symbiotic theory, says that both mitochondria and chloroplasts were once free-living prokaryotes that took up residence inside the large, eukaryotic cell and provided an advantage to the cell-aerobic respiration from mitochondria and photosynthesis from chloroplasts. Over time, the once free-living prokaryotes lost many genes and became unable to live freely. A consensus sequence for related repeated sequences can be derived by examining individual repeats and generating an approximate sequence, having only minor variation among the different repeats. They are often genuine coding sequences that have defects that prevent their expression. Regions that have many adenines tend to move more slowly than regions without adenines. The supercoils can be wound and unwound during processes such as cell division by the action of several enzymes, including gyrase and topoisomerase. Several proteins recognize sequences near to the origin of replication (parS sequences) and help partition the supercoiled chromosome into each new cell during division. Additionally, one adenine pairs with two thymines and one guanine pairs with two cytosines. Histone proteins can also be modified by the addition of acetyl, methyl, and phosphate functional groups, which has a direct effect on gene expression. Nucleosomes are further wound into a 30 nanometer fiber, which can loop back and forth to form either the highly condensed heterochromatin or the more diffuse form, euchromatin. Prior to cell division, the chromatin condenses further in preparation for division. The authors describe a method to visualize chromatin remodeling using fluorescent-based probes that can recognize specific modifications, such as an acetylation of the histone. Additionally, the K12 site was stable, even after removing the inhibitors of histone deacetylation. This phenomena could lead to the discovery of a new mechanism for controlling Brdt stability of specific acetylation events. Overall, the use of these highly sensitive and specific probes to investigate dynamic chromatin modifications could provide the researchers new targets for some drugs and lead to new technologies for evaluating the roles of histone deacetylases and histone acetyltransferases. Do you think that the large, fluorescent proteins affect the structure or acetylation of the chromatin The authors mention that these probes could provide researchers in drug discovery with new targets. Addition of acetyl functional groups to chromatin generates euchromatin, the more diffuse regions of chromatin and potentially, more active forms in terms of gene expression. Why would the reseachers notice a marked deacetylation of K5 and K8 during chromosome compaction That is, why is it beneficial to the cell to remove the acetyl groups at this point in the cell cycle Through the incorporation of the probe, named Histac, the researchers were able to determine that the sites of acetylation were at lysine five (K5) and lysine eight (K8) on histone H4. Both H4 K5 and K8 had to be acetylated in order for Brdt, an acetyl recognition protein domain, to bind the acetylated histone. The Histac probe not only detects acetylation, but it can also detect several other occurrences in the dynamic chromatin. Lastly, Histac detected a loss of hyperacetylation during mitosis as the chromosomes compacted. Researchers using this probe determined that during mitosis, unlike the K5 Manipulation of Nucleic Acids Chapter 1. Small segments can then be purified, analyzed, and rejoined in a variety of configurations. Here, we describe the methods for the synthesis of both natural nucleic acids and some useful chemical derivatives. Finally, the use of nucleic acid hybridization in a variety of situations is introduced to facilitate the further understanding of more modern molecular biology techniques. The first example in this chapter is the discovery of restriction enzymes and other nucleases in bacteria. In particular, the discovery of restriction enzymes that only recognize specific nucleotide sequences has been essential for mapping the location of the various fragments in reference to the other fragments. Some nucleases will only attack single-stranded nucleic acids, others will only attack double-stranded nucleic acids, and a few will attack either kind. Modification enzymes usually add the methyl group to adenine or cytosine within the recognition site, and this group prevents the corresponding restriction enzyme from recognizing the site. Since this sequence is an inverted repeat, the enzyme also cuts the other strand after the corresponding G, giving a staggered cut. Modification enzymes are paired with restriction enzymes and recognize the same sequence. Modification enzymes methylate the recognition sequence, which prevents the restriction enzyme from cutting it. Restriction enzyme recognition sites are usually four, six, or eight bases long and the sequence forms an inverted repeat.
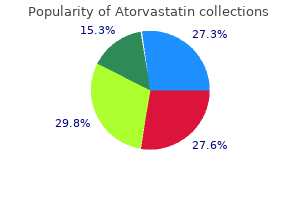
10 mg atorvastatin mastercard
Now do cholesterol lowering foods work 20mg atorvastatin with visa, the histones at the beginning of a gene are physically distinct from the histones toward the center or end of a gene. Describe the role of the following protein modifications on transcription in eukaryotes: a) methylation b) acetylation c) phosphorylation Discussion points 1. What other cases have you come across of repeated segments in proteins and what role do these play Upstream elements lie upstream at moderate distances and bind a range of specific transcription factors. Enhancers also bind specific transcription factors but lie much further away and may be either upstream or downstream. This is also a carefully controlled process, although many of the details are still obscure. These modifications are essential for their proper function in protein translation (see Ch. In fact, certain introns are self-splicing; that is, they cut themselves out in a reaction that does not require any protein components (see below). Initial processing involves ribonucleases that cut the primary transcript at the sites shown by arrows. When translated, only the information from the exons is used to build the protein. If the primary transcript were translated as is, it would result in a huge, dysfunctional protein containing many extra stretches of random amino acids due to the intron regions. The primary transcript is trapped inside the nucleus until it is completely processed. Possible additions of other methyl groups to the ribose of nucleotides #1 and #2 are indicated. This is another indication of the prokaryotic ancestry of these organelles (see Ch. Splicing must be accurate to within a single base since a mistake would throw the whole coding sequence out of register and totally scramble the protein sequences. In the middle of the intron is a special adenine residue used as a branch site during splicing. B) the overall binding of U1 at the 5 splice site and of U2 at the branch site are shown. B) the 5 splice site is cut first and the free end of the intron loops back to bind the branch site. In particular, U6 displaces U1 from the 5 splice site and U1 and U4 are lost from the complex. First, the intron and exon are cut apart at the 5 splice site and the free 5 end of the intron loops around and is joined to the adenine at the branch site. Second, the free 3 end of the upstream exon displaces the intron from the 3 splice site and the two exons are joined together. Different Classes of Intron Show Different Splicing Mechanisms There are several classes of introns (Table 12. They are processed in an almost identical manner, by a different, but closely related, set of splicing factors. The 3D structure is folded so as to bring the two splice sites together and to strain the bonds that will be broken. For clarity, here we have indicated only the first level of intron folding by base pairing. A) In Group I introns, the 5 splice site is attacked by a soluble guanosine nucleotide that cuts the exon and intron apart. Exon 1 Exon 2 Introns are found in Archaea but are removed by simple ribonucleases without needing a splicesome. However, the reaction is started by attack of an internal adenosine (not a free nucleotide as in Group I introns). Twintrons are complex arrangements in which one intron is embedded within another. Since introns are embedded within other introns, they must be spliced out in the correct order, innermost first, rather like dealing with parentheses in algebra. R-Loop Analysis Determines Intron and Exon Boundaries Electron microscopy has been used for direct visualization of eukaryotic introns. B) the entire complex can be visualized by electron microscopy after shadowing with metal ions. Generally, alternative splicing is used by different cell types within the same animal. At first glance it might seem that alternative splicing provides, at least in theory, a way for each gene to encode multiple proteins, hence increasing the total number of different proteins available to an organism. However, selection of which alternative splice site to use in a particular cell or tissue must itself be controlled, and this often requires several additional proteins. Alternative Promoter Selection Alternative promoter selection occurs when two alternative promoters are available. The choice of which promoter to use depends on cell-type specific transcription factors. Alternative Tail Site Selection Alternative tail site selection may occur when alternative sites for adding the poly(A) tail are possible. In this case, cleavage at the earlier poly(A) site results in loss of the distal exon. If the later poly(A) site is chosen, then the earlier poly(A) site and the exon just in front of it are spliced out. This mechanism is used to produce antibodies that recognize the same invading, foreign molecule but that have different rear ends. One type of antibody is secreted into the blood, whereas the other type remains attached to the cell surface. Alternative Splicing by Exon Cassette Selection Alternative splicing by exon cassette selection involves a genuine choice between actual splicing sites. If promoter #1 is used, then the segment containing promoter #2 and exon #2 is spliced out. Transcription yields a single primary transcript that is spliced in two alternative ways. The same primary transcript is drawn twice in different ways to illustrate the two splicing plans. Some cell-type specific factor that recognizes the different possible splice sites must come into play here, but the details are still obscure. Exon cassette selection occurs in the gene for the skeletal muscle protein troponin T. Five (exons 4 through 8) may be used in any combination (including none used) and the final two (exons 17 and 18) are mutually exclusive, and one or the other must be chosen. The result is that muscle tissue contains multiple forms of this structural protein. Trypanosomes are parasitic single-celled eukaryotes that cause sleeping sickness and other tropical diseases. They evade immune surveillance by constantly changing the proteins on their cell surfaces by the genetic trick of shuffling gene parts. On the other hand, trypanosomes do not appear to have introns and so do not have normal splicing! Inteins and Protein Splicing Occasional intervening sequences are found that are spliced out at the protein level. Such protein splicing is rare, which is why it was only noticed relatively recently. In other words, inteins are intervening sequences in proteins that are present when the protein is first made, but are later spliced out. Certain specific amino acids must be present at the extein/intein boundaries for the splicing reaction to work. Serine (or cysteine) must be the first amino acid of the downstream extein, as its hydroxyl group (or sulfhydryl if cysteine is used) is needed to carry the upstream extein during the branched stage. Usually there is just a single intein per protein, but examples are known where multiple inteins are inserted into the same host protein.
Atorvastatin 40 mg line
This early embryo becomes then in its entirety surrounded by a fluid-filled chorionic cavity cholesterol jama buy atorvastatin australia, called also extraembryonic coelom, which develops within the loosely arranged extraembryonic mesoderm. The embryonic disc remains linked to the developing placenta through the connecting stalk. Together with the process of gastrulation, the seemingly simple embryonic disc acquires several morphologically and molecularly defined axes of polarity. The dorsoventral axis of the embryo is defined by the ectoderm facing the amnion dorsally versus the hypoblast facing the yolk sac ventrally. The caudocephalic axis is defined by the primitive streak caudally versus the developing neural plate and oropharyngeal membrane cranially. Subsequent to its formation the intraembryonic mesoderm differentiates into the axial notochord, intermediate mesoderm, and lateral plate mesoderm. Concomitantly, the extracellular spaces between the cells of the lateral plate mesoderm unite to form an intraembryonic coelom. The intraembryonic coelom splits the mesoderm into two layers: the splanchnic mesoderm facing the endoderm and the somatic mesoderm leaned against the ectoderm. Cranially and caudally, small round regions of epiblast and hypoblast are not separated from each other and form the oropharyngeal and cloacal membranes, which are the future entrance and exit of the gastrointestinal tract. Rising the Heart: the Primary Field of Cardiac Progenitors At the cranial part of the trilaminar embryonic disc a part of the splanchnic mesodermal layer consists of cells destined to differentiate into cardiomyocytes. This part of the lateral plate mesoderm is called the cardiogenic mesoderm, and consists of two populations of cardiac progenitors, the so-called heart fields (36,37,38,39). The epithelial sheath of splanchnic mesodermal cells differentiating first and forming the primitive heart tube makes up the primary heart field. The adjacent pool of cardiogenic cells located central and dorsal to the primary heart field and differentiating at later stages makes up the secondary heart field. Although the primary and secondary heart fields share a common origin (40), they contribute cells to the developing heart in a temporal- and spatialspecific manner (41,42). The primary heart field cells are characterized by the expression of several myocardial contractile proteins and "cardiac" transcription factors. Induction of the differentiation of the cardiogenic mesoderm toward the primitive myocardial phenotype occurs by signaling cross talk between multiple tissue types. The transcriptional programs that guide the specification of splanchnic mesoderm into primary myocardium require first the expression of the T-box transcription factor Eomesodermin (46), which at the primitive streak stage directly activates the transcription factor Mesoderm posterior-1 (Mesp1) (47). The induced cardiogenic mesodermal cells only acquire a myocardial phenotype when imposed upon by a balance of positive and negative stimuli emanating from the neighboring endodermal, ectodermal, and notochordal cells. Signaling from endodermal cells positively regulates cardiac specification through multiple transcription factors and signaling pathways including Sonic hedgehog (Shh), Wnt ligands, P. Signals from the neural plate, somatic and axial mesoderm, in turn, repress myocardial differentiation (38), mainly through the Wnt signaling pathway. B: A drawing of a longitudinal cross-section through such an early human embryo showing the embryonic disc surrounded by the amniotic cavity facing the epiblast (blue cells), and the yolk sac facing the hypoblast (yellow cells), which, in turn, are surrounded by the chorionic cavity lined by the villi of the forming placenta. During this process the cells of the epiblast detach from their neighbors, acquire a mesenchymal phenotype, and migrate through the primitive streak "inside" the embryonic disc to form the three embryonic germ layers, endoderm, mesoderm, and ectoderm. Subsequent to the formation of the mesodermal layer, its extracellular spaces merge together to form the intraembryonic coelomic cavity. The intraembryonic coelom divides the lateral plate mesoderm into a somatic layer, facing the ectoderm and a splanchnic layer, facing the endoderm. As a result of folding, the foregut is formed and the bilateral cardiogenic plates are brought together ventral to the foregut, where they eventually fuse to form the primitive heart tube. Note that already at these early stages the paired dorsal aortas are present between the forming neural tube and the foregut. The endothelial cells of the vessels bringing blood to the forming heart tube (future veins), the endocardial cells of the heart tube, and the endothelial cells of the vessels bringing blood away from the heart tube (future aortic arches and dorsal aortas), are initially morphologically identical. In the mouse while the gap junctional protein connexin 37 is expressed in all endothelial cells, connexin 40 only is expressed in the endothelial cells of the arteries, but not in the endocardium or veins (56). The endocardial cells covering the initially bilateral lumens of the forming heart tube are derived from the same cardiogenic mesodermal cells as the primary myocardium. In the heart-forming region differentiation of the endothelial cells is induced by local signals from the endoderm and the cardiogenic mesenchyme to form bilateral endothelial tubes that will fuse to form the midline endocardial tube. The initial step of endocardium formation is delamination of the endothelial precursor cells from the precardiac mesoderm. Angiopoietins are known to stimulate endothelial precursor cell migration and proliferation. Gata5 is transiently expressed and largely restricted to the endocardial cells in the cardiogenic mesenchyme. Gata5 is induced concomitant with Flt1 and prior to induction of Tie2, ErbB3, and connexin 37 in endothelial differentiation. As the endocardial cells become terminally differentiated, several factors are upregulated including epicardin, endothelin-1, tenascin X, and endothelial-specific transcription factor Epas1 (65,66,67). A: Shows a ventral view of a very early mouse embryo at the stage of formation of the crescentic primary field of heart progenitors, as visualized here by expression of myosin light chain (dark blue color). Box in (C) details the molecular interactions leading to the formation of the cardiogenic mesoderm. The negative and positive influences of the endodermal, notochordal, and ectodermal tissues are depicted by red lines and green arrows, respectively. The future endocardial cells (depicted by yellow), adjacent to the endodermal layer (blue), separate early from the cardiogenic mesenchyme (rose), destined to differentiate into myocardium. Under influence of endodermal signals (blue arrow) and of angiopoietin secreted by cardiogenic mesenchyme (red arrow), the proendocardial cells undergo further differentiation and acquire definitive endocardial phenotype (red). Such vascular precursors then unite into vessels which become continuous with the vessels of the connecting stalk (68,69). The molecular mechanisms underlying the formation of the first embryonic veins and arteries are not known. It is unclear how the mesodermal cells at exactly determined locations are programmed to differentiate into endothelial cells and to arrange themselves in circular fashion in continuity with one another along the longitudinal axis of the embryo. It is also not known how the lumens within these very first vessels are formed, albeit the morphologic changes are quite well described (70,71). Formation of the Primitive Heart Tube Concomitant with the differentiation of the primary heart field the embryo acquires its characteristic curved shape by the process of folding of the initially flat embryonic disc (72). Because the peripheral margins of the embryonic disc hardly grow in contrast to the disc itself, the forming embryo bulges out over the margins, which gives the embryo its shape. By this process also the lateral parts of the embryo are brought together ventrally at the midline. Largely due to the enormous growth of the developing brain and surrounding tissues, the embryo folds in longitudinal direction, by which the forming heart tube and its surrounding coelomic (future pericardial) cavity rotate 180 degrees along the transverse axis and become located ventrally and caudally to the developing brain. During the lateral folding of the embryo, the myocardial sheets of splanchnic mesoderm steadily bulge into the coelomic cavity. Subsequently they fuse together, forming the bowl-shaped primary cardiac tube being bordered dorsally by the foregut and filled with a gelatinous substance called cardiac jelly. The bowl-shaped myocardial structure continues to bulge into the pericardial cavity until its lateral edges meet dorsally at the midline and fuse to complete the formation of the heart tube. The fusion of the bilateral heart-forming regions and subsequent formation of the embryonic bowlshaped heart have been demonstrated to depend on the contractile properties of the adjacent endodermal layer (74). The linear heart tube initially remains attached to the dorsal wall of the pericardial cavity by the dorsal mesocardium, which disappears during the process of cardiac looping at later stages, by which the heart tube acquires an S-shape form. The heart tube itself remains attached to the coelomic wall at its arterial and venous poles. Initially, it has been thought that looping of the heart tube simply is the consequence of rapid growth of the tube within a much more slowly expanding pericardial cavity.
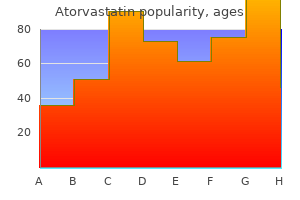
Buy cheap atorvastatin 5mg
Although the picture on how these proteins function is becoming clearer cholesterol avocado buy discount atorvastatin, much is still unknown. But when these proteins do not function properly, bacteria are unable to properly express their genes and die, suggesting that the organization is essential for the bacteria to survive. This is the sum of the contributions due to the double helix plus the supercoiling. In this terminology, the linking number, L, is the sum of the twist plus the writhe (L T W). As each supertwist is introduced, gyrase changes conformation to an inactive form. When one strand is broken, the other strand is passed through the break to undo one supercoil. Behind the moving replication and transcription apparatus, a corresponding wave of negative supercoiling is generated. As a result, at any given instant, the extent of supercoiling varies greatly in any particular region of the chromosome. Local supercoiling depends largely on the balance between transcription and the restoration of normal supercoiling by gyrase and topoisomerase. In practice, the four to eight base sequences recognized by most regulatory proteins and restriction enzymes are too short to yield stable cruciform structures. Their existence can be demonstrated because they allow single strand specific nucleases to cut the double helix. The basic idea of electrophoresis is to separate the molecules based on their intrinsic electrical charge. Electrically-positive charges attract negative charges and repel other positive charges. Conversely, negative charges attract positive charges and repel other negative charges. Two electrodes, one positive and the other negative, are connected to a high voltage source. Positively-charged molecules move towards the negative electrode and negatively charged molecules move towards the positive electrode. The larger molecules find it more difficult to squeeze through the gaps in contrast to the small pieces that can wiggle through the meshwork with ease. When agarose and water are mixed and boiled, the agarose melts into a homogeneous solution. As the solution cools, it gels to form a meshwork, which has small pores or openings filled with water. The cooled gel looks much like a very concentrated mixture of gelatin without the food coloring or flavoring. The pore size of agarose is suitable for separating nucleic acid polymers consisting of several hundred nucleotides or longer. Agarose gels are normally square slabs that allow multiple samples to be run side by side. The more superhelical twists, the more compact the molecule is and the faster it moves in an electrophoretic field. Watson and Crick (Nature (1953) 171: 737) described the most stable and most common of these. The grooves running down the helix are different in depth and referred to as the major and minor grooves. The A-form of the double helix is shorter and fatter than the B-form and has 11 base pairs per helical turn. In the A-form, the bases tilt away from the axis, the minor groove becomes broader and shallower, and the major groove becomes narrower and deeper. B) Actual ethidium bromide stained electrophoresis gels of four differing concentrations (A: 6. Note that the sequence of each individual strand is written in the 5- to 3-direction. Its existence can be demonstrated in small plasmids by changes in electrophoretic mobility. It has been suggested that regions of Z-helix may be specifically recognized by certain enzymes. Since the chicken is diploid, there are two chromatin territories for each chromosome. The model proposed by this paper suggests that the chromatin territory is separated from another by an interchromatin space. This organization could partly explain how some eukaryotic genes are expressed and others are not expressed. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. This reduces the repulsion between the three strands and helps form a triple helix. Not surprisingly, the presence of artificial triplexes has been shown to block transcription. The structure of the nucleus at interphase was originally thought to be an amorphous mess of chromatin. Eukaryotic chromosomes may be as much as a centimeter long and must be folded up to fit into the cell nucleus, which is five microns across. Eight histones comprise the core unit: two of H2A, two of H2B, two of H3, and two of H4. The entire length of nucleosomes is called chromatin and because of its appearance, it is also referred to as "beads on a string". The core histones, H2A, H2B, H3, and H4, are small, roughly spherical proteins with 102 to 135 amino acids. The central part of H1 binds to its own nucleosome and the two arms bind to the nucleosomes on either side. The core histones have a body of about 80 amino acids and a tail of 20 amino acids at the N-terminal end. This tail contains several lysine residues that may have acetyl groups added or removed. In the second level of compaction, the chain of nucleosomes is wound into a giant helical structure with six nucleosomes per turn known as the 30 nanometer fiber. The nucleosomes may form a tubular solenoid shape, or they may zigzag back and forth. Only one of the H2A and H2B dimers is shown; the other is on the other side, hidden from view. The 30 nm fibers are further folded by looping and attachment to a protein scaffold. C) the 30 nm fibers form loops that are periodically anchored to protein scaffolding. Just prior to mitosis, condensation occurs, making the chromosomes (and chromatids) visible. The chromosomes are best viewed while spread out during the middle part (metaphase) of mitosis. Each daughter cell will acquire one of the chromatids and the process begins anew. About 10% of this euchromatin is even less condensed and is either being transcribed or is accessible for transcription in the near future (see Ch. Note that between cell divisions and in non-dividing cells each chromosome consists of only a single chromatid. The term chromatid is mostly needed only to avoid ambiguity when describing chromosomes in process of division. In addition, there are a few unusual cases where giant chromosomes with multiple chromatids are found in certain organisms. This associated preview article discusses how two new probes are being used to ascertain whether or not histone H4 is acetylated on lysine 5 (K5), lysine 8 (K8), and lysine 12 (K12). The group of Minoru Yoshida created a genetic fusion of the bromodomain gene, Brdt, and the gene for histone H4 connected by a linker region for flexibility.
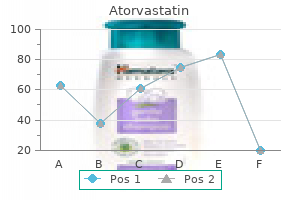
Order atorvastatin 5 mg with visa
The control sites and initiation points are present on the expression vector for both transcription and translation high cholesterol medical definition purchase atorvastatin once a day. The result is the protein of interest fused to a tag that can be used to purify and detect the protein. Polyhistidine-tagged proteins are purified using a chromatography column containing divalent cations, usually nickel (Ni2). In addition to short peptide tags, whole length protein tags can be fused to a protein of interest. Fusion proteins with Protein A tags are purified using antibodies against Protein A. Before the protein of interest becomes functional, the fusion tag must be removed by cleavage with a proteinase. The use of self-splicing inteins as purification tags eliminates the need for proteinases to remove the tags. Protein Interactions and the Interactome l Phage display allows genes to be isolated by detecting the proteins they encode. In phage display, a bacteriophage is used to carry both the gene of interest and the protein product. Protein-protein interactions are detected in phage display by construction of phage display libraries. The total amount of protein-protein interactions occurring within a cell is called the protein interactome by some individuals. The trophoblast is the outer layer of the blastocyst and provides nutrients to the embryo and becomes a portion of the placenta. Through their method, the authors generated cleaner and higher yield samples during the purification process. In all, 92 proteins, excluding Oct4 itself, were identified as Oct4-associated proteins. The identifications of some of the proteins were confirmed by Western blot analysis. Thirteen of the 92 proteins were previously characterized as Oct4 interacting proteins. Additionally, eight previously identified Oct4-interacting proteins were not detected or were detected within the controls. The authors performed tandem affinity purification on the tagged Oct4 and revealed seven more proteins. Since these proteins bound to Oct4 through more stringent purification, the authors believed they represent the highest-affinity proteins associated with Oct4. Over 50% of the Oct4-associated proteins have been confirmed by independent means, confirming that the data set produced by these authors is representative of the Oct4 binding partners. Why must the nucleotide sequences for protein tags be cloned in frame to the target protein Discussion points Using the new purification tag, the authors were able to purify Oct4 plus its binding partners under specific purification conditions. How might changing the conditions of the purification change the outcome of the experiment Do you think the authors could have missed some of the binding proteins for Oct4 simply because of the purification method Hence, transcription will not occur unless interaction of the two proteins occurs. In co-immunoprecipitation, the gene for the protein of interest is expressed in animal cells and isolated using antibodies either to the protein itself or to a fusion tag. Any proteins intracellularly associated with the protein of interest are also purified as long as the isolation conditions are conducive for such interactions. Protein Arrays l Protein arrays are built using tagged proteins and are screened by a variety of approaches. Protein Interactions and the Interactome e313 Protein arrays are used for the global analysis of proteins within cells. Analysis of proteins includes the biochemical and enzymatic analysis as well as protein-protein interactions. These arrays are constructed by tagging proteins with chemical groups (for example, by genetic engineering of their genes) which are then used to anchor the fusion proteins to a solid surface. These bound proteins are then assayed for enzymatic activity or protein interactions with the help of an easily detectable substance, such as a fluorescent dye. Protein arrays use fusion tagged proteins for immobilization of the protein on the solid surface. Metabolomics provides a means to analyze differences in mutant strains of bacteria and growth conditions by identifying different metabolic intermediates present within the samples. Conceptual questions the field of functional genomics attempts to assign function to an unknown gene whose physiological role is not well-understood. However, protein sequence similarity does not necessarily translate into functional similarity. The authors used a recombinant protein with unknown function and incubated it with cellular extract from Mtb. How does the field of metabolomics change the assignment of putative functions to unknown gene products Discussion points In this paper, the researchers used a metabolomics approach to reassign the function of an Mtb enzyme based upon the metabolic intermediates. Do you think that changing the growth conditions could affect the outcome of their experiment Although some genes are expressed all the time, most are expressed only under conditions where they are needed. Transcription is the first step in gene expression and, not surprisingly, is the site of a great deal of regulation, which is the topic of the present chapter. However, the mere presence of a protein is not sufficient to ensure a correct physiological response and the activity of many proteins is regulated after they are made, as discussed in Chapter 14. Gene Regulation Ensures a Physiological Response In bacteria such as Escherichia coli, about 1,000 of the 4,000 genes are expressed at any given time. Three gels were run for each condition and the figure shows a layered view of two three-gel composites. The pink and green spots are proteins induced in acetate or formate, respectively. The circled spots are those that were statistically validated, based on a pair-wise comparison of all the individual gels. Single-celled organisms regulate their genes in response both to changes in the environment (such as temperature, osmotic pressure, or availability of nutrients) and to the internal state of the cell (such as readiness for cell division). In multicellular organisms, one must also consider communication between cells and the developmental progression of the organism as a whole. Any of these individual steps may be regulated, but in practice, regulation of certain steps is much more common than the regulation of others. For example, the initiation of transcription is more often controlled than its elongation phase. For an efficient response-in other words, one requiring less energy-the gene is often regulated at the level of transcription. When a rapid response is needed, a precursor protein is produced in advance and then rapidly converted to an active protein when the conditions warrant. If a rapid response is critical to the survival and propagation of a species, making an inactive enzyme and holding it on standby is a good strategy. Bacteria must respond not only to the "natural" environment, but those that inhabit animals or plants must also be prepared to counter the attempts of their host organisms to eliminate them (see Focus on Relevant Research). In general, regulation of transcription is most efficient for the organism, but control of enzyme activity allows for a fast response. In positive regulation, a gene is incapable of expression unless it receives a positive signal of some sort. Some genes are regulated positively, others negatively, and still others by multiple regulators, including both types. There are many different diseases that are caused by bacteria, and although antibiotics have changed the way most infections are treated, many bacteria are resistant to antibiotics. Although some bacteria cause disease, a large number are actually beneficial to humans. The intestine is filled with a large number of different bacteria that aid digestion of different foods. Because of the long-standing relationship between humans and bacteria, humans have developed elaborate immune systems to fight off harmful bacteria and nourish good bacteria.
Cheap atorvastatin 5mg visa
This is crucial in the neonate to ensure the transition to extrauterine life cholesterol lowering foods fish order atorvastatin online from canada, postnatal cardiac growth, and to facilitate -adrenergic signaling prior to the arrival of sympathetic nerve terminals (170). Conversely hypothyroidism correlates with bradycardia and mild hypertension (171). Promising results have been obtained in several small-scale clinical studies using T3 therapy in infants and children undergoing cardiopulmonary bypass, showing that this therapy improves postoperative cardiac function (173,174). In summary, the combination of neurohormonal programming and autonomic innervation and mechanical forces coordinate postnatal cardiac growth and E-C coupling in order to increase cardiac output to meet the metabolic demand of the rapidly growing organism. Pioneering work in the late 19th century by Frank (177) and in the early 20th century by Starling et al. As described earlier, this response is mediated by an increased responsiveness of the myofilament apparatus to Ca2+ at longer sarcomere lengths (158). This allows ventricular filling and involves the interaction between active, energy-dependent processes (myocardial relaxation) that primarily influence early ventricular filling and passive processes, such as loading conditions and myocardial compliance. Alterations in Pump Function at Birth Alterations in hemodynamic load during gestation and after birth can impact cardiac function. These processes seemingly prepare fetuses for the cardiac and systemic autonomy that occurs at birth. Unquestionably, the process of birth is a significant physiologic strain on both the mother and offspring. The switch from fetal circulation to the complete separation of oxygen-rich and oxygen-depleted blood in the neonatal period is a consequence of the physiologic orchestration of decreased pulmonary vascular resistance, increased systemic vascular resistance, ductus arteriosus and ductus venosus closure, coupled with the closure of the foramen ovale in the heart. Using newer speckle tracking echocardiography technology, this study also revealed significantly decreased longitudinal strain and strain rate and increased myocardial velocities in the neonatal period compared to the fetal period. Collectively, these data indicate that the fetal heart chambers adapt differentially in response to the unique changes in hemodynamic load that occurs at birth to maintain systemic perfusion. These powerful diagnostic tools have both strengths and limitations depending on their research and clinical utility. As relaxation continues, the pressure decreases but volume remains constant since the mitral valve has not yet opened (line E-A). Ca2+ binds to troponin C, causing conformational changes of troponin I (TnI) and troponin T, which removes Tm and allows for myosin binding to the actin thin filaments. Although this systolic relationship is generally considered nonlinear, a linear line can be fit to this curve as long as the volume axis intercept is also considered. Emerging Concepts and Conclusion Research into the developing myocardium is still in its infancy, with numerous studies emerging within the last 5 years. The role of cardiac-resident stem cells in the prenatal and postnatal heart remains a popular area of study. New concepts concerning the epigenetic regulation of cardiac maturation, especially in the intrauterine and neonatal stages have emerged as important focus of clinical and translational research. The clinical impact of these studies is particularly pertinent since the effects of prematurity and maternal stresses (preeclampsia, obesity, gestational diabetes) on postnatal heart structure and function are largely unknown. These areas of research will likely continue to increase our understanding of the developing heart and how it functions, and assist in developing novel therapeutic approaches for the treatment of pediatric cardiovascular diseases. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Three-dimensional architecture of cardiomyocytes and connective tissue in human heart revealed by scanning electron microscopy. Interactive relationship between basement-membrane development and sarcomerogenesis in single cardiomyocytes. Tissue distribution of the laminin beta1 and beta2 chain during embryonic and fetal human development. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Developmental changes in the ultrastructure and sarcomere shortening of the isolated rabbit ventricular myocyte. Human fetal heart development after mid-term: morphometry and ultrastructural study. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. Gene regulation, alternative splicing, and posttranslational modification of troponin subunits in cardiac development and adaptation: a focused review. Rapid purification of mammalian cardiac troponin T and its isoform switching in rat hearts during development. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Dissociation of structural and functional phenotypes in cardiac myosin-binding protein C conditional knockout mice. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stresssensing and mechanical dysfunction. Calcium sensitivity and the Frank-Starling mechanism of the heart are increased in titin N2B region-deficient mice. Formation of highly organized intracellular structure and energy metabolism in cardiac muscle cells during postnatal development of rat heart. Hypoxia signaling controls postnatal changes in cardiac mitochondrial morphology and function. Mechanotransduction: the role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function. Mechanical stress-induced sarcomere assembly for cardiac muscle growth in length and width. Restoration of resting sarcomere length after uniaxial static strain is regulated by protein kinase Cepsilon and focal adhesion kinase. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Current understanding and management of dilated cardiomyopathy in Duchenne and Becker muscular dystrophy. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. The neonatal heart has a relatively high content of total collagen and type I collagen, a condition that may explain the less compliant state. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Phosphorylation of connexin43 on Serine368 by protein kinase C regulates gap junctional communication. N-cadherin is required for the differentiation and initial myofibrillogenesis of chick cardiomyocytes. Assembly of the cardiac intercalated disk during pre- and postnatal development of the human heart. Electrophysiological effects of remodeling cardiac gap junctions and cell size: experimental and model studies of normal cardiac growth. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries.
Buy atorvastatin 10 mg mastercard
There are several groups that are distinct enough that some scientists would elevate them in rank to miniature kingdoms how many cholesterol in shrimp buy atorvastatin 5mg amex. Once thought to be plants that had lost their chloroplasts, it is now thought they never had them. Like plants, fungi are nonmobile, but they lack cellulose and their cell walls are made of chitin. After an organism is classified into a kingdom, then each kingdom is divided into phyla (singular, phylum). Genera are divided into species, such as Homo sapiens Biological classification attempts to impose a convenient filing system upon organisms related by continuous evolutionary branching. Some Widely-Studied Organisms Serve as Models With so much biological diversity, biologists concentrate their attention on certain living organisms, either because they are convenient to study or are of practical importance, and they are termed "model organisms. Nonetheless, information discovered in such model systems is assumed to apply also to related organisms. But since they do have limitations; ultimately, human cells and agriculturally-useful animals and plants have to be studied directly. Bacteria Are Used for Fundamental Studies of Cell Function Most of the early experiments providing the basis for modern-day molecular biology were performed using bacteria such as E. Furthermore, a bacterial culture consists of many identical cells due to lack of sexual recombination during cell division. In contrast, in multicellular organisms, even an individual tissue or organ contains many different cell types. All the cells in a bacterial culture respond in a reasonably similar way, whereas those from a higher organism will give a variety of responses, making analysis much more difficult. The most commonly used bacteria have about 4,000 genes as opposed to higher organisms, which have up to 50,000. Furthermore, different selections of genes are expressed in the different cell types of a single multicellular organism. Bacteria are haploid, having only a single copy of most genes, whereas higher organisms are diploid, possessing at least two copies of each gene. Analyzing genes present in a single copy is far easier than trying to analyze two different alleles of the same gene simultaneously. The bacterial population may reach 5 109 cells per ml or more in only a few hours under ideal conditions. Plasmids have been used extensively by scientists as vectors to carry different genes from different organisms. Bacteria can be grown under strictly controlled conditions and many will grow in a chemically-defined culture medium containing mineral salts and a simple organic nutrient such as glucose. Bacteria grow fast and may divide in as little as 20 minutes, whereas higher organisms often take days or years for each generation. Consequently, genetic experiments that need to analyze large numbers of cells can be done conveniently. Thus, it is not necessary to keep hundreds of cultures of bacterial mutants constantly growing just to keep them alive. In practice, bacteria are usually cultured by growing them as a suspension in liquid inside tubes, flasks, or bottles. They can also be grown as colonies (visible clusters of cells) on the surface of an agar layer in flat dishes known as Petri dishes. Agar is a carbohydrate polymer extracted from seaweed that sets, or solidifies, like gelatin. It should be noted that the convenient properties noted above apply to commonly-grown laboratory bacteria. In contrast, many bacterial species found in the wild are difficult or, by present techniques impossible, to culture in the laboratory. Many others have specialized growth requirements and most rarely grow to the density observed with the bacteria favored by laboratory researchers. The biological diversity of bacteria is immense since the total number of bacteria on our planet is estimated at an unbelievable 5 1030. The total amount of bacterial carbon is 5 1017 grams, nearly equal to the total amount of carbon found in plants. Probably over half of the living matter on Earth is microbial, yet we are still unable to identify the majority of these species. The bacteria that live in extreme environments, such as boiling hot water in the deep sea thermal vents, salty seas like the Dead Sea, and Antarctic lakes that thaw for only a few months each year, have very unique adaptations to the Biologists have always been pulled in two directions. The research on these bacteria will provide a wealth of information of how life formed, exists, and persists. For example, in 1997 the Hudson Foods plant in Columbus, Nebraska was forced to shut down and 25 million pounds of ground beef were recalled. These are chemical substances capable of killing most bacteria by inhibiting specific biochemical processes, but which are relatively harmless to people. The most commonly used antibiotics, the penicillins and cephalosporins, are synthesized by a kind of fungus known as mold. However, many antibiotics are made by one kind of bacteria in order to kill other types of bacteria. The Streptomyces group of soil bacteria produces a wide range of antibiotics including streptomycin, kanamycin, and neomycin. Some antibiotics, like chloramphenicol, were originally made by molds, but nowadays can be chemically synthesized. Finally, some antibiotics, such as sulfonamides, are entirely artificial and are only synthesized by chemical corporations. When penicillin is produced by molds grown on agar in a Petri dish, it will diffuse outwards and suppress the growth of bacteria in a circle around it. Its natural habitat is the colon (hence "coli"), the lower part of the large intestine of mammals, including humans. In addition, bacteria, together with their viruses and plasmids, have been used experimentally during the genetic analysis of higher organisms. Outside the cytoplasmic membrane possessed by all cells are the cell wall and a second, outer membrane. However, it can be inconvenient to the biotechnologist who wishes to manufacture genetically-engineered proteins from genes cloned into E. Consequently, there has been a recent upsurge of interest in gram-positive bacteria, such as Bacillus, which lack the outer membrane. In 1946, Joshua Lederberg was attempting to carry out genetic crosses with bacteria. Until then, no mechanisms for gene transfer had been demonstrated in bacteria, and genetic crosses were therefore thought to be restricted to higher organisms. Because the plasmid carries the genes for fertility, it was named the F-plasmid (see Ch. Various genes are indicated with numbers corresponding to their position on the map. The replication origin (oriC) and termini (ter) of replication are also indicated. Note that chromosome replication does not start at zero map units- the zero point is an arbitrary designation. Yeast Is a Widely-Studied Single-Celled Eukaryote Yeast is widely used in molecular biology for many of the same reasons as bacteria. It is the eukaryote about which the most is known and the first whose genome was sequenced-in 1996. Yeasts are members of the fungus kingdom and are slightly more related to animals than plants. Even before the age of molecular biology, yeast was widely used as a source of material for biochemical analysis. The first enzymatic reactions were characterized in extracts of yeast, and the word enzyme is derived from the Greek for "in yeast. Although larger than bacteria, yeast cells are only about a tenth the size of the cells of higher animals. The natural life cycle of yeast alternates between a diploid phase and a haploid phase.
Purchase discount atorvastatin on-line
However boost good cholesterol foods cheap atorvastatin 20mg mastercard, the recent massive increase in sequence data has shown that most virus proteins have no known homologs among present-day cellular proteins. This implies that much viral genetic information originated either during the replication of viruses themselves or that it derives from cellular lineages that are now extinct. Some specifically viral proteins, including capsid proteins, are shared among viruses that infect host cells from all three domains of life. This argues that some viruses are extremely ancient and that their ancestors predated the Last Universal Common Ancestor of cellular life forms (see Ch. The origin of viruses and, in particular, the invention of the virus particle as a means for moving between host cells is still a mystery. Typically, satellite viruses infect and hence damage the same host cell as their helper virus. Instead of replicating separately in the host cell they invade and infect the virus assembly compartment where mimivirus replicates. This, paradoxically, benefits the host cell since the virophages are tiny relative to the colossal mimivirus (50 nm versus 750 nm; 20 genes versus 1000). They reduce algal death from phycodnaviruses and overall promote growth of the algae in the marine environment. As explained in Chapter 23, "Mutations and Repair," this is related to their high mutation rates. Since each mutant gene product must continue to interact with other virus components, this limits the number of genes to no more than a dozen or so. The advantage of a high mutation rate to these viruses is that it allows them to change their proteins rapidly so evading recognition by host defense systems. The terms positive and negative refer to the coding strand and the non-coding strand, respectively. The sense strand can also be referred to as the plus strand and the antisense strand as the minus strand. Conversely, if the virus particle contains the minus strand, it must first make the complementary, positive strand before moving on to manufacture proteins. He observed that milkmaids who worked in close contact with cattle often caught cowpox, a mild disease. After deliberately infecting patients with cowpox he found that they had indeed become immune to smallpox. Jenner called the material he used "vaccine" after the cows where it originated (vacca cow in Latin) and the novel technique was named vaccination. For a long time it was believed that cowpox virus and the vaccine virus were identical. However, in 1939 it was found that cowpox virus (re-isolated from cows) and the virus cultures maintained for use in vaccination were actually quite distinct. Since viruses could not be cultured or purified in the laboratory until much later, those who followed Jenner constantly re-isolated new strains of what they thought was cowpox virus from cows and horses. At some time the original cowpox was replaced by a different virus, the present vaccinia virus. No virus presently circulating in the wild corresponds to vaccinia so its precise origin is unknown. Furthermore, the differences between vaccinia and cowpox are too large for mutation to be responsible for cowpox evolving into vaccinia. Presumably another poxvirus was circulating among cattle and horses during the nineteenth century and was eventually kept because it is even milder than cowpox. Although the genome is very small, the three proteins are all that the virus needs to successfully infect bacteria and replicate itself. Not only does the virus avoid using capdependent translation itself, but it shuts it down, thus preventing host protein synthesis (see Focus on Relevant Research). Because picornaviruses use an internal ribosome entry site for viral protein synthesis they can shut down host-cell protein synthesis by suppressing cap dependent translation. Polyproteins are made from a single giant gene and then cut up to give several final proteins. These viruses vary in shape and structure but are similar in having an outer envelope derived from the membrane of the host cell where they were assembled. The plus strand is used as a template to manufacture more negative strands for the next generation of virus particles. The name "mosaic" refers to the yellowish blotches on the leaves of infected plants. X-ray crystallography and other physical methods were used to show that cylindrical viruses are in fact made of helically-arranged protein subunits. Therefore, viruses that use this type of genome must synthesize the complementary plus strand upon entry into the host cell. This illustrates that the capsid proteins are held together largely by the hydrophobic force (see Ch. This membrane layer has retrovirus proteins both inserted through it and covering its surface. Consequently, retroviruses are impossible to get rid of completely after infection and integration, at least using current medications and procedures. Like many other animal viruses, the retrovirus has an outer membrane derived from the host cell. This envelope contains viral proteins that coat the entire surface of the virus particle. Some of the proteins are embedded in the outer envelope while others sit upon the surface. As with bacteriophage lambda, this first gives a defective virus, in which host genes have replaced virus genes. However, recombination with a wild-type retrovirus can generate a functional virus that carries host genes as well as a complete retrovirus genome. Oncogenes are genes that are involved in regulating cell division in animals and when mutated cause cancer. The oncogenes carried by viruses are originally of animal origin and were picked up by the virus from some previous host animal. Although these cancer-causing viruses have attracted most notice, in principle any host gene close to the site of integration could be moved by retrovirus transduction. It is especially useful in obtaining copies of eukaryotic genes that lack the non-coding intervening sequences. Genome of the Retrovirus A typical retrovirus has three major genes: gag, pol, and env. Many of the genes overlap in sequence with each other and are shown side-by-side in the diagram. The protein products of gag, pol, and env are actually polyproteins that give rise to two or three proteins after cleavage. The products of the three major genes, gag, pol, and env, are polyproteins that are cleaved to generate several shorter proteins (Table 21. The vast majority of such sequences are defective and many have large internal deletions. From one to several thousand copies of the genomes of each type of these retroviruses may be present. Based on sequence comparison, most endogenous retroviruses occupied their present sites before divergence of humans and other primates. A few have entered the human genome since the human/chimpanzee split (approximately 5 million years ago). These are all in the same place in different human groups, implying that new additions happen very infrequently and that once integrated the endogenous retroviruses rarely move from one position to another. For example, virus particles related to endogenous retrovirus sequences have been isolated from patients with multiple sclerosis, but whether these contribute to the disease or are a side effect of tissue damage is unknown. There is also evidence that some human promoters, enhancers, and alternative splice sites may have been recruited from the remains of endogenous retroviruses. These may be self-replicating but lack a protein coat or they may be defective and depend on a helper virus to provide replication genes and/or a protein coat. Such satellite viruses are only capable of replication and/or packaging when another virus, the helper virus, is present to provide the necessary gene products.