Generic 5 mg aricept amex
This study found that in all type B strains examined treatment quotes images buy aricept with paypal, the tpeL gene was always encoded 3 kb after the cpb gene. The cpb plasmids of type B strains co-exist with at least one other virulence plasmid, an epsilon toxin (etx)-encoding plasmid, and sometimes another accessory toxin plasmid, encoding lambda toxin (lam) and a putative urease operon (44). In these strains the size of the etxencoding plasmid and other accessory plasmids is consistent, with the only variable plasmid being the cpb-encoding plasmid (44). The limited variation of these plasmid combinations may be due to incompatibility issues with resident plasmids present in type B isolates. No type B plasmids have been identified that encode both the cpb and etx genes (44). In contrast, the cpb-encoding plasmids of type C strains demonstrate much greater size variation (45). In these strains, the beta toxin gene is encoded on plasmids ranging in size from 65 kb to 110 kb (45). Restriction analysis suggests that some type C isolates may carry the same cpb plasmids found in type B strains (90- and 65-kb plasmids) (45). However, in addition to size differences, the cpb-encoding plasmids of type C strains also differ in their carriage of accessory toxin genes (45). As in type B strains, cpb is often associated with tpeL (45), although tpeL and cpb are not always encoded on the same plasmid (45). It is the third most potent bacterial toxin, after tetanus and botulinum toxins (47). Activation of epsilon toxin can be facilitated by proteolytic cleavage catalyzed by the C. Once active, epsilon toxin primarily targets endothelial cells near the intestinal border, forming a prepore complex before inserting itself into cell membrane regions following binding to specific cellular receptors (53, 54). The toxin oligomerizes, forming a pore in the cell wall and inducing cellular damage (55). These damaged endothelial cells then allow epsilon toxin to be absorbed, where it systemically targets organs such as the lungs, and kidneys and in the brain, where it induces the release of the excitatory neurotransmitter, glutamate (47, 56, 57). Excessive glutamate release and subsequent overstimulation of neurons results in altered neurological capabilities, which are commonly observed in animals suffering from epsilon toxin-induced enterotoxemia (47, 57, 58). The etx gene is harbored on conjugative plasmids found in both type B and D strains of C. The etx plasmids of type B strains again appear to have a limited degree of plasmid diversity, with the etx gene only encoded on an 65-kb plasmid (44). In contrast, the etx plasmids of type D strains are more variable in size, ranging from 45 to 110 kb (60). In addition to the size variability observed for type D etx plasmids, they are also more diverse in their toxin gene carriage (60). Some of the larger etx plasmids have been found to encode numerous other toxin genes such as cpe, lam, and cpb2, which encodes beta2 toxin (60). Whether the toxin genotype of a type D strain is simple (carrying only plc and etx) or more complex (possessing other toxin genes such as cpe or cpb2) appears to delineate the size of the plasmid on which the etx gene is located (44). It comprises two protein components: Ia -(the enzymatic component) and Ib -(the binding component) (62, 63). Each component alone is nontoxic, but when combined the intact toxin produces rapid cytotoxic effects that often result in death (64, 65). These genes are located on large virulence plasmid that range in size from 65 kb to 135 kb (66, 67). Many type E strains carry additional genes, such as those encoding for urease and lambda toxin, on their iota plasmid (66). The components of iota toxin, like epsilon toxin, are activated by proteolytic processing (64), and the lambda toxin protease is known to process iota toxin into an active form (64, 69). The carriage of both toxin genes on a single plasmid would be considered advantageous because it would be beneficial for cells expressing iota toxin to also encode its activator, lambda toxin (66). In all strains characterized both lam and iap/ibp were encoded on the same large conjugative plasmid (66). These complexes then form pores in the cell membrane, activating cell death pathways that result in cellular destruction (71). It is encoded by the cpe gene, which can be located both chromosomally and on conjugative plasmids (74, 75), which appears to play a role in the epidemiology of disease (75, 76). It is thought that the carriage of cpe on conjugative plasmids is a key factor in the disease phenotype observed during nonfood born gastrointestinal infections (40). The cpe gene is encoded on numerous plasmids that range in size from 70 kb up to 110 kb (45, 60, 66, 77). Plasmids encoding cpe have been found in type C, D, E and F strains, but not in any type B isolates (45, 60, 66, 77). However, the sequence downstream of cpe differs greatly, depending on the different genetic locations (78). The cpe plasmids show significant diversity and can differ in their carriage of other toxin genes (45, 60). In some type C strains cpe can either be carried on the same plasmid as cpb or on a separate plasmid (45). The larger etx-encoding plasmids characterized in type D strains tend also to carry cpe genes (60). NetB is a b-pore forming toxin which is the causative agent of necrotic enteritis in chickens and forms the basis for the new toxinotype G. Sequencing of these strains identified a putative toxin which was denoted NetB, Necrotic Enteritis Toxin B-like (80). Deletion of netB resulted in the formation of a strain that was unable to produce necrotic lesions in chickens (80). Complementation with the netB gene in trans restored the strain to wild-type virulence (80). The beta2 toxin gene, cpb2, is possibly the most promiscuous of all toxin genes, because it can be found on plasmids from all C. Despite its name, beta2 toxin has little amino acid sequence identity (less than 15%) to beta toxin (84). The involvement of beta2 toxin in disease is often debated because there is no direct evidence of beta2 toxin-induced virulence (85). The cpb2 gene is carried by numerous plasmids ranging in size from 45 to 90 kb (44, 45, 60, 81, 82). These plasmids can carry numerous other toxin genes such as cpe and etx, with one plasmid characterized in a type D strain carrying all three toxin genes (60). In many strains cpb2 is carried on a conjugative plasmid distinct from other toxin plasmids, as is seen in many type B to G strains (44, 45, 66, 81, 82). Although it has not been shown to be involved in disease, it is thought that there may be a synergistic effect between delta and beta toxins, because they are often produced by the same strains (88, 89). Delta toxin is produced primarily by type C strains and possibly some type B strains (89, 90). Sequencing of the strains identified three genes-netE, netF, and netG- that were predicted to encode pore forming toxins with 59. Virulence Plasmids of the Pathogenic Clostridia 959 sequence similarity to NetB (91). In the first netE/F/Gpositive type A strain characterized, both netE and netF were found to be encoded on the same large conjugative plasmid (92, 93), whereas netG was located on a second large conjugative plasmid that also carried the cpe gene (91). Sequencing of other netF-positive strains showed that cocarriage of netE and netF on the same plasmid remained consistent; however, carriage of the netG plasmid was variable among these isolates (92, 93). There is also a strong association of netF-positive strains isolated from diseased dogs and foals which, along with the cytotoxicity assays, suggests that NetF is involved in these diseases (91). Further genetic characterization of netF-positive strains has demonstrated that netE/netF and netG are encoded on two distinct and relatively large pathogenicity loci on their respective plasmids (92, 93). However, due to the presence of TpeL in some hypervirulent type G avian necrotic enteritis strains it has been suggested that TpeL may play a synergistic role with other toxins such as NetB (95).
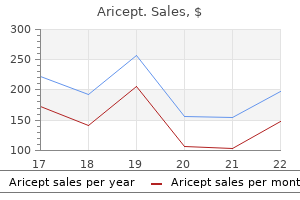
Discount aricept line
Influence of cranberry proanthocyanidins on formation of biofilms by Streptococcus mutans on saliva-coated apatitic surface and on dental caries development in vivo medications that cause hair loss discount aricept 5 mg online. The tea catechin epigallocatechin gallate suppresses cariogenic virulence factors of Streptococcus mutans. Targeted killing of Streptococcus mutans by a pheromone-guided "smart" antimicrobial peptide. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Targeted antimicrobial treatment to re-establish a healthy microbial flora for long-term protection. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. YlxM is a newly identified accessory protein that influences the function of signal recognition particle pathway components in Streptococcus mutans. As a consequence, beneficial aspects of streptococcal colonization of the oral cavity were initially overlooked. The first comprehensive analysis of the resident oral microbiota was accomplished in 2005 (1), and with this, a new picture began to emerge. With the availability of highthroughput sequencing techniques and an increased sensitivity in analysis methods, the presence of a defined microbiome associated with oral health has been shown (2). Alongside this, "omics" techniques have revealed that prevalent oral diseases such as caries and periodontal disease are polymicrobial in nature and the result of microbial dysbiosis (3, 4). Even more striking, the metabolic output of these mixed microbial communities seems to be more relevant than their precise microbial composition (4). This is also reflected by the fact that the severity of caries and periodontal disease is heavily influenced by the synergistic interactions of the individual members of the polymicrobial consortium, including metabolic cross-feeding and interspecies signaling with transcriptional adjustment to the metabolic output. Thus, the ecological context of the microbial community seems to be of importance to understanding oral health and disease development. Novel approaches to combat oral polymicrobial diseases should therefore focus on the bacterial community that is present in the healthy oral cavity. Since oral streptococci are abundant during initial colonization of the tooth (8, 9), their function is to provide a favorable environment for incorporation of later species and to support accretion of the mature oral biofilm, which in general has a health-protecting function (10, 11). A more recent analysis using the housekeeping genes rpoB, sodA, ddl, and gdh showed a more distant relationship with S. First, the ecological niche is occupied, which plays a significant role in the process of colonization resistance and the exclusion of potential incoming pathogens. Second, initial colonization shapes the overall composition of the biofilm, since the pioneer colonizer can produce metabolic products or provide a substratum for compatible partner species. Complex formation is facilitated through electrostatic interactions of salivary proteins with the tooth surface to form the acquired enamel pellicle, a process that occurs within seconds after a clean enamel surface is exposed to saliva (21). Microbial attachment to the acquired enamel pellicle is then mediated via protein-protein doi:10. The protein content of the acquired enamel pellicle is thus a major determinant of colonization sequence, dictating the strength of microbial adhesion as well as localization, since pellicle proteins seem to differ according to anatomical site (22). A prominent protein in saliva and the acquired enamel pellicle is a-amylase, which is responsible for the catalytic hydrolysis of starch (23, 24). Amylase-binding proteins have been identified in several oral streptococcal species (24). Mutation of AbpA results in deficient biofilm formation and bacterial adhesion in vitro (26). This function has been shown to be mediated by long filamentous pili organized in a four-gene operon (ssa1631 to ssa1634) that encodes three pilin subunits (PilA-C) and a dedicated sortase (SrtC) for cell-surface anchoring of the pilin structural proteins (28). This poses a potentially interesting dynamic with AbpA, which is strictly confined to the outer cell surface, as shown for S. Taking into account that pili are flexible, one could hypothesize that binding to amylase in acquired enamel pellicle is possible even when the molecule is scarce, since the pili could serve as a flexible "arm," latching onto free amylase within saliva. A recent publication demonstrating that amylase in acquired enamel pellicle is actually less abundant than that in saliva (29) is in agreement with this hypothesis, suggesting an advantage of a flexible arm over a rigid arrangement on the bacterial surface. The pili also showed binding to other salivary proteins, and their deletion diminished biofilm formation on saliva-coated surfaces (28). However, the mutant was still able to bind amylase, albeit with lower efficiency (28), suggesting that other surface proteins are also able to bind amylase, possibly the aforementioned AbpA homolog (27). A pilus-bound amylase also offers the advantage of retaining about 50% of its enzymatic function (30). Gaining access to the amylase substrate starch through a flexible pilus would increase the chance for 28. Genetics of sanguinis-Group Streptococci in Health and Disease 451 hydrolysis of the alpha-1,4-glycosidic linkage into glucose, maltose, and maltodextrins, promoting sugar uptake and subsequent metabolism by simply increasing the accessible radius of the cell. The majority of mucins are synthesized and secreted by the submandibular and sublingual glands, as well as minor glands located in the palatal, buccal, and labial mucosae. Mucins are heavily glycosylated glycoproteins and form a lubricating, viscoelastic coating on all oral surfaces. SrpA contains a subdomain in its binding region that is similar to the V-set Ig-like fold adopted by mammalian Siglecs (sialic acidbinding immunoglobulin-like lectins) (34, 35). Indeed, Siglec-like domains have been identified in potential adhesins of several S. Glycoarray dot blots with human salivary samples and naturally occurring glycoconjugates have demonstrated a high specificity of S. The glycoarray also revealed an interesting role for divalent cations Ca2+ and Mg2+ in the binding of S. This is further supported by the increased abundance of proteins known to bind divalent cations such as Ca2+ in the acquired enamel pellicle compared to saliva (29). SsaB is able to bind to saliva-coated hydroxyapatite through an unknown mechanism (37), but its principal function seems to be the transport of divalent cations (38). While this transport was demonstrated to be specific for Mn2+ and Fe2+, playing a pivotal role in oxidative stress defense (38), other divalent cations such as Ca2+ and Mg2+ might still be able to interact with SsaB to facilitate binding to the acquired enamel pellicle. Spreading zones after prolonged incubation can be observed around colonies, indicating active movement (39). Whether or not this so-called twitching motility has any function or is used as a means to disseminate within the oral biofilm is not yet clear. This correlates with the observation that signal peptidase I is required for biofilm development (40). In general, signal peptidases are membrane-bound endo-proteases that cleave the signal peptide portion from the majority of secreted proteins (41). Although the substrates for signal peptidase processing are not experimentally verified, in silico prediction identified 168 potential candidates, including several adhesins (40). Further characterization of signal peptidase-processed surface proteins will most likely identify new proteins involved in the adhesion process. This process then leads to a series of ordered and temporal events, in which coadhesion predominates, ultimately resulting in formation of a mature biofilm.
Diseases
- Hereditary ceroid lipofuscinosis
- Penoscrotal transposition
- Retinopathy aplastic anemia neurological abnormalities
- Sosby syndrome
- Histadelia
- Proctitis
- Short stature mental retardation eye defects
- Lung neoplasm
- Proteus syndrome
Cheap aricept 10 mg mastercard
The final layer of the cell envelope is the capsule symptoms underactive thyroid buy 10mg aricept with mastercard, mainly consisting of polysaccharides and proteins. The cell envelope is highly impermeable and unique to other Gram-positive bacteria. The additional membrane components SecD, SecF, and YajC increase the efficiency of export. The list of SecA2-dependent exported proteins includes examples with and without a signal peptide. Although the mycosin protease (MycP) is not an integral component of the secretion complex, it associates with the complex and is essential for successful secretion. It is currently unknown how Sec, SecA2, Tat, and T7S substrates are secreted across the outer membrane into the capsular layer or culture supernatant. Protein Export across the Mycobacterial Cell Envelope 1131 the mechanism for crossing the outer membrane remains a mystery. In all bacteria, the majority of this protein export is carried out by the general Sec pathway (16). Mycobacteria have 1,000 predicted Sec-exported proteins (this number includes proteins exported across as well as into the inner membrane), many of which have been experimentally demonstrated to be exported (17, 18). Among the many proteins exported by the Sec pathway are proteins with essential functions in the cell envelope. Further, there are Sec-exported proteins with critical roles in host-pathogen interactions, making the Sec pathway critical for bacterial pathogenesis as well (17, 18, 20). The Sec pathway is highly conserved in bacteria, and most of our understanding of this pathway comes from studies of the Escherichia coli system (for recent reviews of the Sec system see references 21 and 22). Mycobacteria possess conserved orthologues of all the critical Sec pathway components, and as expected, these Sec orthologues are either proven or predicted by saturating mutagenesis transposon-site hybridization or transposon insertion sequencing studies to be essential (23). While only a few mycobacterial Sec proteins have been directly studied, the results so far indicate that the Sec pathway of mycobacteria functions similarly to the Sec pathways of other bacteria. Sec-exported proteins can remain in the cell envelope or through subsequent mechanisms transit across the additional cell envelope layers and be fully secreted. Sec-exported proteins are in an unfolded state during their translocation through the channel (25). There are also additional membrane components that improve Sec export efficiency: SecD, SecF, and YajC (29). For mycobacteria, the second step a Sec-exported protein may take to cross the outer membrane remains completely unknown. Consistent with SecA1 being the SecA of the essential general Sec pathway, SecA1 of Mycobacterium smegmatis and M. Following protein synthesis, Sec-exported proteins exist in the cyto- plasm as unfolded preproteins with N-terminal signal peptides that are 25 to 30 amino acids long (38). The Sec signal peptide is composed of a positively charged N-terminus, a hydrophobic central domain, and an uncharged polar C-terminus containing a signal peptidase cleavage site (39). For lipoproteins exported by the Sec pathway, there is also a lipobox motif at the C-terminus of the signal peptide with an invariant cysteine that serves as the site of lipid attachment and anchoring of the protein to the cell envelope postexport (40). In association with export, the signal peptide is removed from the preprotein by one of two signal peptidases (LepB for the majority of Sec preproteins and LspA for lipoprotein preproteins specifically), which serves to produce the cleaved, mature protein species on the extracytoplasmic side of the membrane (39). The dispensability of LspA is also the case for Gram-positive Firmicutes, while in Gram-negative bacteria, LspA is essential (44). The general principles of signal peptides established for other bacteria also apply to mycobacterial proteins exported by the Sec pathway, as shown by in silico prediction programs for Sec signal peptides being good predictors of M. In addition to possessing a signal peptide, another required feature of Sec-exported proteins is that they must be unfolded to be exported (25, 46, 47). Specifically, for membrane proteins with large periplasmic domains, SecA may assist in exporting these domains across the membrane (56). Similar to the Sec pathway, after transport from the cytoplasm, Tatexported proteins can either remain in the cell envelope or be fully secreted via a subsequent mechanism. Proteins exported by the Tat pathway are synthesized as preproteins with cleavable N-terminal signal peptides. However, as the name of the pathway reflects, Tat signal peptides are distinguished from Sec signal peptides by a Tat motif containing a pair of twin arginine residues (see below) (62). Compared to the Sec pathway, fewer total proteins are exported by the Tat pathway. Some Streptomyces strains are estimated to have 150 proteins exported by the Tat pathway, and M. Most notably, the Tat pathway is fundamentally different from the Sec pathway in that folded, rather than unfolded, proteins are exported (66). The studies conducted so far of the mechanism of Tat export in mycobacteria indicate that the Tat system of mycobacteria functions similarly to the Tat pathways of other bacteria. In Firmicutes, however, the Tat pathway is a streamlined system with only TatA and TatC (65). Tat preproteins are recognized by a complex of TatB and TatC, with TatC recognizing the twin arginine signal peptide (71, 72). Tat export is a posttranslational process, and the energy required to drive Tat transport comes from the proton motive force (73, 74). Some models propose that TatA oligomers form a substrate-fitted protein-conducting channel (75) that accommodates folded proteins of different sizes and shapes. Following export, Tat preproteins undergo signal peptide cleavage by the same signal peptidases that work with the Sec pathway: LepB or LspA (77, 78). Tat signal peptides resemble Sec signal peptides in having a positively charged N-terminus, a hydrophobic central domain, and an uncharged polar C-terminus containing a cleavage site. However, Tat signal peptides are distinguished from Sec signal peptides by the presence of a Tat motif with twin arginines (R-R-X-F-F [F = hydrophobic]) (62). In mycobacteria, Tat signal peptides appear to follow the same rules established in other bacteria. Since b-lactamases must be exported to protect bacteria against b-lactam antibiotics, the requirement of the Tat pathway for BlaC export contributes to the natural resistance of M. In addition to a Tat signal peptide, the second required feature of Tat-dependent proteins is that they are folded prior to export. Unfolded or misfolded proteins are generally unacceptable for export, suggesting that the Tat export system has a "proofreading" ability that restricts export to properly folded proteins (66, 82). In addition to fully exporting proteins across the membrane, the Tat pathway plays a role in the delivery of a small subset of integral membrane proteins into the inner membrane. Most recently, examples of integral membrane proteins that require both the Sec and Tat pathways to deliver and transport across the membrane multiple periplasmic domains were identified. The role of the Tat pathway in this interesting subset of proteins is to translocate periplasmic domains that, similar to many soluble Tat substrates, must bind a cofactor in the cytoplasm and, therefore, fold prior to export. The first example of a dual Sec- and Tat-dependent membrane protein was the Streptomyces Rieske iron-sulfur binding protein, which is a subunit of the cytochrome bc1 complex (85). In general, the Tat pathway is not essential for bacterial viability, and tat mutants are viable, at least in standard laboratory media (65). It is important to note that not all SecA2 systems are similar or evolutionarily conserved (35, 95). These systems are called multisubstrate SecA2 systems or SecA2-only systems, the latter name referring to their lack of a second SecY (SecY2) in the system. Staphylococcus and Streptococci are examples of bacteria with SecA2 systems that include an accessory SecY (SecY2) and additional accessory Sec proteins (98). These latter systems export a single large glycosylated substrate, and they are called accessory Sec (aSec) or SecA2-SecY2 systems and will not be discussed further here. We expect there are distinctive features of SecA2 substrates and additional components of the SecA2 pathway that distinguish it from the general Sec pathway.
Purchase aricept 10mg on-line
A recent study led to the design medicine 4212 buy aricept 5mg lowest price, synthesis, and characterization of a GlcN6P analogue, carba-GlcN6P, which constitutively activates the glmS ribozyme of vancomycin-resistant S. Very recently, a new approach called "Term-seq" revealed that several antibiotic resistance genes are under the control of riboswitches responding to antibiotics commonly used against Gram-positive pathogenic bacteria such as Listeria spp. Not surprisingly, the presence of antibiotics can also modulate the regulatory activity of T-boxes. Thereafter, the T-box senses the availability of glycine not only for its incorporation into nascent polypeptide chains during translation but also for the formation of pentaglycine bridges into the peptidoglycan molecule, linking two essential pathways. Based on sequence and structure conservation, most of the T-boxes and riboswitches were predicted in S. The outcome depended on the binding sites of the protein synthesis inhibitors (11). The best-studied example in Gram-positive bacteria is the thermosensor regulating the expression of the transcriptional factor prfA, which activates the expression of most of the virulence genes in Listeria monocytogenes at high temperatures (13, 14). No such example has yet been demonstrated for other Gram-positive bacteria, including S. Such elements can potentially be transferred horizontally to other bacterial species or be duplicated (20). Initially, pervasive transcription was considered a nonfunctional transcriptional noise. The situation might be even more complex, because recent results suggested that the termination factor Rho plays a major role in preventing pervasive transcription in Bacillus subtilis, but also in S. Three genes were transcribed from the positive strand and two from the negative strand. Moreover, one small open reading frame was detected within one of the genes from the minus strand and coded for a secreted peptide with similarity to the RelE toxin (32). Interestingly, this locus was expressed in a growthphase-dependent manner, in nutriment starvation, and in oxidative stress. In several cases, a second distinct site of interaction occurs in the coding region. Indeed, there is no identifiable proQ homolog, and the role of Hfq is still unclear. However, while the SarA level remains relatively constant during bacterial growth, the expression of SprC fluctuates, which suggests that additional factors might control its synthesis and that a mechanism of derepression should coexist under specific conditions. Red lines corresponded to posttranscriptional regulation, and black lines, to transcriptional regulation. Outnumbering their microbial hosts, phages impose selective pressure for the diversification of microbial defense systems. During a second infection event they are transcribed and used as guides to inactivate the viral or plasmid genome. However, the acquisition step involves two highly conserved Cas1 and Cas2 proteins (58, 63). Different endonucleases participate in the maturation step, which might vary in different bacteria. The peculiarity of this system (SaCas9) is that it contains a shorter version of Cas9 protein. The discovery of the smaller Cas9 protein led to recent improvements in genome editing (81). Interestingly, SaCas9 has been successfully used for eukaryotic genome editing since its smaller size makes it easier to be delivered via adeno-associated virus vectors to somatic tissues (81). This is often accompanied by the expression of various virulence factors involved in the colonization and the alteration of the tissue but also by the capacity to escape from the host immune response. For instance, it represses the synthesis of protein A, which triggers inflammatory signaling pathways and contributes to evasion of the immune response. Recent modeling of the quorum sensing system and of its regulators has illustrated the importance of the agr system in promoting dissemination of the bacteria from biofilms or dense populations (93, 94). Perhaps more surprisingly, heterogeneity has been reported in patients in which agr-positive and agr-negative strains coexisted. This has been proposed as a factor that might modulate the outcome of the infections (100, 101). Some of these genomic islands play key roles in pathogenesis through their possession of new virulence factors (pathogenicity islands) or through the synthesis of novel regulators modulating the expression of genes of the core genome. However, the phenotype of the DsprD mutant strain was not linked to the SprD-dependent regulation of sbi since the Dsbi mutant strain behaved like the wild-type strain in the mouse sepsis model (102). Therefore, these data strongly suggest that SprD might regulate the expression of other proteins important for infection. Indeed, the virulence of the isogenic strain lacking SprC was significantly and reproducibly enhanced in a mouse systemic model. This activation is linked to a specific binding of the phosphorylated form of AgrA to the promoter sequences upstream of the psm genes, except that this binding was not yet demonstrated for the psm-mec gene (reviewed in 110). This deletion is linked to the deregulation of MgrA, a master regulator of capsule synthesis and clumping (112). This phenotype in pathogenesis is probably linked to the high sensitivity of the mutant strain to opsonophagocytosis by host polymorphonuclear leukocytes. Moreover, biological variables may influence the analysis since the relationships between host immune systems and microbes seem to be particularly individualized and can influence the disease outcome (113, 114). Furthermore, interspecies interactions between bacterial pathogens and the commensal microbiota, as well as limited nutrients, play major roles in promoting or preventing S. Interestingly, it was shown that the agr system is repressed by high concentrations of hemoglobin in the nasal fluids, leading to the expression of several cell surface proteins and favoring nasal colonization (115). Bidnenko V, Nicolas P, Grylak-Mielnicka A, Delumeau O, Auger S, Aucouturier A, Guerin C, Repoila F, Bardowski J, Aymerich S, Bidnenko E. Termination factor Rho: from the control of pervasive transcription to cell fate determination in Bacillus subtilis. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. Stamatopoulou V, Apostolidi M, Li S, Lamprinou K, Papakyriakou A, Zhang J, Stathopoulos C. Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors. Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, Vandenesch F, Caldelari I, Romby P. Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, Geissmann T, Boisset S, Romby P, Vandenesch F. Tomasini A, Moreau K, Chicher J, Geissmann T, Vandenesch F, Romby P, Marzi S, Caldelari I. Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. Nitzan M, Fechter P, Peer A, Altuvia Y, Bronesky D, Vandenesch F, Romby P, Biham O, Margalit H. A semi-quantitative model of quorum-sensing in Staphylococcus aureus, approved by microarray meta-analyses and tested by mutation studies. Growth phasedependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Song J, Lays C, Vandenesch F, Benito Y, Bes M, Chu Y, Lina G, Romby P, Geissmann T, Boisset S. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factor H and C3b. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K.
Discount aricept 5mg free shipping
As one would anticipate medicine for vertigo generic aricept 10 mg amex, the solution of the X-ray structures of DtxR and the ternary complex that forms with its binding to toxO confirms and extends the earlier observation that its footprint compasses a region of 30 bp immediately upstream of the transcription initiation signal (46). While the overall mechanism of DtxR binding to toxO is similar to that of other prokaryotic repressors, there are some unique interactions that should be noted. Each helix-turn-helix in the dimer makes a total of nine interactions with backbone phosphate groups. Since there is a structural rearrangement of activated DtxR upon binding to the tox operator, Thr7 in helix A also interacts with a backbone phosphate group, and Ser37 and Pro39 contact methyl groups on thymine bases in the tox operator through van der Waals interactions (47). The second metal ionbinding site, or ancillary site, is composed of five residues: His79, His98, Glu83, Glu170, and Gln173 (41, 43, 49). The role played by the ancillary metal ion-binding site was elucidated through the analysis of DtxR(E175K), a hyperactive mutant that remained active in vivo even in the presence of the chelator 2,2dipyridyl (50). Nuclear magnetic resonance solution structures combined with other biophysical studies have suggested that apo-DtxR exists in a partially unstructured molten globule, which upon coordination with divalent transition metal ions undergoes a structural conversion to a discrete ordered tertiary structure that both dimerizes and is able to bind to the tox operator (52). The resulting conformational changes then allow the binding of the second metal ion to the ancillary site and subsequent dimerization of DtxR and the formation of an active repressor (53). Under reducing and denaturing conditions, nicked diphtheria toxin may be separated into an enzymatically active N-terminal 24-kDa fragment A and its 38-kDa C-terminal fragment B (54, 55). These preloaded red cell ghosts were then fused to diphtheria toxinresistant mouse L-cells by Sendai virus. Using a fluorescenceactivated cell sorter, L-cells that fused with a single red cell ghost were then isolated and grown for 7 days. Careful analysis of the colony-forming ability of the recipient cells compared to the control cells clearly demonstrated that the delivery of a single molecule of fragment A to the cytosol was sufficient to kill that cell. As such, it was realized through biochemical and genetic analyses that native diphtheria toxin was a protein with at least three structural-functional domains: (i) catalytic, (ii) transmembrane or translocation, and (iii) receptor-binding domains. The toxin is readily purified from the spent culture supernatant by ammonium sulfate precipitation followed by ion exchange chromatography on a diethylaminoethyl matrix. While the intact toxin is enzymatically inactive, exposure of purified toxin to trypsin or other serine proteases 69. Diphtheria Toxin and the tox Operon 1159 proved to be correct with the determination of the crystal structure of diphtheria toxin (60, 61). The N-terminal catalytic domain consists of amino acids Gly1 to Cys186 and is composed of eight b-strands that form two b-sheets forming a core structure that is surrounded by seven short a-helices. The catalytic domain is connected to the centrally positioned translocation domain by a 14-amino acid loop that is subtended by a disulfide bond between Cys186 and Cys201. This exposed loop carries an ArgArgValArg protease recognition site for either furin or other trypsin-like proteases. The proteolytic cleavage of this site is essential for the release of the catalytic domain into the eukaryotic cell cytosol (62). The translocation domain encompasses amino acids Cys186 to K385 and is composed of nine a-helices and their connecting loops. This channel is formed by translocation helices 5 to 7 and 8 to 9, which are highly hydrophobic and form two membrane-soluble "daggers" (61). The loops connecting translocation helices 8 to 9 and 5 to 6 are highly acidic and carry a total of six Asp and Glu residues. The above-described studies demonstrate that diphtheria toxin has evolved to become an extremely efficient nanomachine in which all of its structural domains play an essential role in the intoxication of sensitive eukaryotic cells. While colonization of a susceptible individual with a toxigenic strain certainly plays a role in pathogenesis, Pappenheimer and Murphy (66) demonstrated that transmission of toxigenicity may also occur by in situ lysogenic conversion of an autochthonous nontoxigenic strain of C. It is remarkable that immunization with diphtheria toxoid results in the production of neutralizing antibodies that not only block diphtheria toxin from binding to its cell surface receptor, but also protects against clinical disease (67, 68). It is well known that mass immunization of populations with diphtheria toxoid has led to a dramatic decrease in the incidence of clinical disease (69). For example, in 1958, before mass immunization with diphtheria toxoid in Romania, only 60% of the population was immune to diphtheria toxin, whereas by 1979, the percentage dramatically increased to 97%. The consequence of either stopping or dramatically reducing a mass immunization program against diphtheria is also known. With the breakup of the Soviet Union, public health immunization programs were dramatically affected and by 1990 only 68% of Russian children had received the appropriate vaccination regimen (71). From the onset of the diphtheria outbreak in 1990 through 1999, more than 157,000 cases and 5,000 deaths were reported. In addition, these investigators also analyzed the nucleotide sequence of the dtxR gene. Fifteen strains in this collection were found to carry two missense mutations in DtxR, A147V and L214I in the C-terminal region of the repressor. Interestingly, the dtxR alleles in the two strains that expressed the highest levels of diphtheria toxin were not found to carry a nucleotide substitution. While most of these mutations did not result in an amino acid substitution, two strains from the Otchakov ribotype carried a G393R mutation in the native receptor-binding domain of the toxin. Of these, the A147V mutation is characteristic of the epidemic clonal group SanktPeterburg/Rossija, which was the dominant strain isolated in the Russian diphtheria epidemic in the 1990s. It is remarkable that the amino acid sequence of diphtheria toxin encoded by the omega-phage in the ParkWilliams strain of C. This conservation of amino acid sequence for at least 100 years suggests that the evolution of new tox alleles that might encode a toxin sufficiently different to avoid neutralization by antitoxoid antibodies is highly unlikely. In the case of this metal ion-activated repressor, the N-terminal region of dtxR was found to carry several missense mutations that did not give rise to an amino acid substitution. These results clearly demonstrated that fragment B translocation domain sequences were required to facilitate the delivery of fragment A, the catalytic domain, into the eukaryotic cell cytosol. The hypothesis was that by choosing a surrogate receptor-binding domain that bound to a cell surface receptor that was both limited in distribution and was internalized into an endosomal compartment that became acidified, it might be possible to develop a family of fusion protein toxins that could potentially serve as a new class of targeted toxins for the treatment of human diseases. Moreover, we reasoned that, if successful, these genetically engineered proteins would constitute a unique family of reagents to deplete discrete subsets of cells and to study the molecular process by which the diphtheria toxin catalytic domain was delivered to the cytosol of targeted eukaryotic cells. While this fusion protein was expressed and secreted into the periplasmic space of recombinant E. Since that time, more than a dozen diphtheria-based fusion protein toxins have been constructed and characterized using amino acid 388 or 389 as the junction between diphtheria toxin and surrogate receptor-binding domain polypeptides. In addition to its effective use in T cell malignancies, Ontak has also been successfully used in the treatment of steroid-resistant graft-versus-host disease. In these instances, Ontak was shown to transiently deplete T regulatory cells and, as such, apparently break tolerance and allow for a more robust host-mounted T-effector cell antitumor response. During the refolding process, it was necessary to add Tween20 to partially prevent the formation of insoluble drug aggregates and allow for refolding of the drug into a biologically active conformation. Furthermore, we modified the gene by adding back the native tox signal sequence so that the fusion protein toxin would be secreted into the culture medium. Finally, we introduced mutations into the downstream half of the palindromic tox operator so that expression was constitutive even in medium with high iron concentrations. Physical mapping of betaconverting and gamma-nonconverting corynebacteriophage genomes. Restriction endonuclease map of the nontoxigenic corynephage gamma c and its relationship to the toxigenic corynephage beta c. Integration of corynebacteriophages beta tox+, omega tox+, and gamma tox- into two attachment sites on the Corynebacterium diphtheriae chromosome. Isolation from Corynebacterium diphtheriae C7(beta) of bacterial mutants that produce toxin in medium with excess iron. Isolation and partial characterization of a corynebacteriophage beta, tox operator constitutive-like mutant lysogen of Corynebacterium diphtheriae.

Generic aricept 5 mg with visa
PrgY has been proposed to function by specifically binding to cognate pheromone as it emerges from the membrane symptoms jet lag cheap aricept master card, either following or concomitant with cleavage by Eep. The protein then either degrades the peptide directly or chaperones it to a cell wall bound protease for degradation (147). TraB/PrgY proteins appear to be part of an orthologous family of proteins found in all domains of life but only spottily distributed (188). For instance, there are representatives in humans, plants, and Gram-negative bacteria but not in any other Firmicutes species outside of the enterococci. This phylogenetic distribution suggests that horizontal gene transfer has been critical for their distribution. No function has been assigned for any of the TraB/PrgY orthologs, but homology suggests that they are involved in some form of peptide signaling or processing. Altogether then, pheromone plasmids encode three proteins, TraC/PrgZ, TraA/PrgX, and TraB/PrgY, that bind their cognate pheromone yet are divergent enough to be able to selectively respond only to the pheromone specific for their encoding plasmid. Structural analyses of these proteins suggest that the pheromone binding pockets did not diverge from a single ancestral domain but rather evolved independently and convergently (148). Assuming that all pheromone plasmids descended from a single ancestor, divergence to respond to new pheromone and inhibitor peptides requires coordinated evolution of the binding pockets of all three proteins. The surface exclusion proteins limit conjugation between strains harboring the same pheromone plasmid (193, 194). Plasmidspecific single-strand nicking by the putative relaxase was demonstrated within a large, conserved inverted repeat in oriT. Specificity was imparted by nonconserved direct repeats, presumed to be the relaxase binding site, adjacent to the conserved inverted repeat (216). The repA/ prgW genes belong to the RepA N family of replication initiator proteins, which along with their associated partition modules, have been recently reviewed (143). RepA N initiator proteins are not restricted to pheromone plasmids and are widespread among conjugative and nonconjugative plasmids in the Firmicutes. The mechanisms of strand opening, initiation of replication, and regulation of initiation frequency have yet to be determined for the pheromoneresponsive plasmids. Both domains flanking the central repeat region of these proteins adopt a structural fold similar to the DnaD primosome. The C-terminal domain retains primosome function and recruits DnaG primase to initiate replication. Furthermore, the Nterminal domain was also demonstrated to be capable of bridging two plasmid origins, suggesting that it could function in replication control by handcuffing (231). It had been previously proposed that the pattern of conservation of RepA N proteins, with a highly conserved N-terminal domain and genus-specific conservation of the C-terminal domain, suggested a universally conserved function. The pheromone-responsive plasmid partition systems are variants of the previously described type Ib class (233). The stipled box within repA/prgW represents the origin of replication (oriV) to which the repA/prgW product binds. The lined boxes at each end of the repB/prgP-repC/prgO operon represent the likely centromere-like sites to which the repC/prgO products bind and, with the repB/prgP product, direct plasmid partition. The fst gene is designated by the diagonally lined box, and the sequence of the peptide is shown at the bottom. Overlapping transcription at the bidirectional transcriptional terminator also provides complementarity. The RepB/PrgP and RepC/PrgO proteins correspond to type Ib ParA and ParB proteins, respectively. A series of similar repeats is also located downstream of the repC/prgO gene (143, 150). The molecular basis of this phenomenon is unknown but may not be directly related to the normal function of the repeats. RepB proteins are broadly distributed in the Firmicutes but are not phylogenetically congruent with the associated replication initiator proteins, suggesting mosaicism of partition and replication modules (143). It consists of a central hydrophobic region flanked by charged N- and Cterminal tails. In all four species overexpression results in condensation and mislocalization of the nucleoid and interference with proper partition and cell division. Internal expression of Fst and external addition of the lantibiotic nisin synergistically effect cell killing, suggesting that the two peptides have different but complementary effects on the cell envelope (252). Saturation mutagenesis experiments revealed that the central hydrophobic region and charged residues in the N-terminus were important for overexpression-mediated toxicity but that the C-terminal charged tail was nonessential (251). An atomic resolution structure determined in a membrane mimetic by nuclear magnetic resonance spectroscopy revealed that the essential hydrophobic region forms an a-helix that spans the membrane with the charged N- and C-termini protruding (253). The charged C-terminal seven amino acids were disordered and predicted to extend from the cytoplasmic side of the membrane. The authors suggested that the primary function of membrane insertion was to facilitate interactions with a specific target, rather than being directed against the membrane itself. They also predicted that the disordered C-terminus might become structured upon recognition of the target. This would appear to contradict results indicating that the C-terminal amino acids are nonessential for toxicity, but it should be noted that the expression system used in reference 251 could not distinguish levels of toxicity. Therefore, it is possible that the toxicity is substantially decreased in the truncated mutants and could even be an artifact of overexpression. These toxins have been divided into eight superfamilies, and Fst is the founding member of the Fst/Ldr family (255). Stems, loops, and bulges are depicted in their approximate locations and sizes as determined by experimentation. Once complex formation is complete (D), the structure is extremely stable in vivo and in vitro perhaps due to the gap between the interacting repeats. The Ldr members are present in the g-proteobacteria and are regulated in a manner more similar to hok-sok family members. In spite of their phylogenetic distance, Ldr overexpression has effects on nucleoid condensation and cell division similar to those Fst has in E. It was found that, while there was some overlap, overexpression of the two toxins had distinct transcriptomic effects (258). This suggests that the toxins have different targets or have distinct effects on the same target. Finally, overexpression of the Fst proteins is approximately 10-fold more toxic to mannitol-grown than to glucose-grown cells (K. Weaver, unpublished observation), strengthening the circumstantial link between par function and carbon flux. The mutant strain showed increased virulence and improved survival in macrophages under several stress conditions. These differences could be strainrelated, due to differences between transcription and translation effects, or due to the accumulation of compensatory mutations in the deletion strain. Two Tn3-type transposons, Tn917 encoding erythromycin resistance and Tn1546 encoding vancomycin resistance, have been extensively studied (260, 261), and Tn917 and its derivatives have been used for transposon mutagenesis in a number of Gram-positive organisms. Many of these elements contain antibiotic resistance genes, and some are conjugative by means that have yet to be fully explained (see below). Conjugative transposons were later found to be widespread in the lactic acid bacteria, and mechanistically similar but unrelated elements were identified in Bacteroides species (266). Two transposonencoded proteins, Int and Xis, are required for excision (276, 277). The N-terminal domain (Int-N) binds via a three-stranded b sheet to direct repeat sequences near each end of Tn916 (280, 281). The C-terminal domain (Int-C) binds to the transposon termini as well as to target sequences. Like other tyrosine recombinases, Int produces staggered cuts at each end of the integrated transposon. One cut is made flush with the end of the transposon, producing a recessed 3 end covalently jointed to Int via a phosphotyrosine linkage.
Chinese Prickly Ash. Aricept.
- Pain, vomiting, diarrhea, abdominal pain, snakebite, skin diseases, and other conditions.
- Are there safety concerns?
- What is Chinese Prickly Ash?
- Are there any interactions with medications?
- How does Chinese Prickly Ash work?
- Dosing considerations for Chinese Prickly Ash.
Source: http://www.rxlist.com/script/main/art.asp?articlekey=97065
Order aricept online now
Notably medications 1 gram buy aricept online pills, the skin of atopic dermatitis patients is often permanently colonized by S. In general, the nasal microbiome is less dense and less diverse than, for instance, the gut microbiome (31, 32). Whereas Staphylococcus auricularis is part of the external ear microbiome, Staphylococcus saprophyticus often colonizes the rectum and genitourinary tract (29). Staphylococcus lugdunensis is also part of the human skin flora and is particularly prevalent in the pelvic and perineal regions, the groin, the axillae, and in the nail bed of the first toe. Attempts to distinguish persistent from intermediate carriers by a single nasal swab indicated that the detection of more than 103 S. Staphylococcus aureus Colonization of the Human Nose 725 defined by pseudostratified, columnar ciliated epithelial cells (2, 41). In the anterior vestibule, the moist squamous epithelium on the septum adjacent to the nasal ostium harbors the highest number of S. Human nasal secretions contain sodium chloride at concentrations found also in other body fluids and small amounts of potassium, magnesium, and phosphate, but they are low in potential nutrients such as sugars, amino acids, and other major building blocks (30). It is likely that secretory proteases produced by a specific bacterial strain generate peptides and amino acids that can be utilized by many other microbiome members and may have a broad impact on microbiome metabolism. IsdA, which is involved in heme uptake and iron acquisition (54), is expressed during human infections and is responsible for nasal colonization and survival of S. Similar to ClfB, IsdA interacts with cytokeratin 10 and loricrin, as well as with the extracellular matrix protein involucrin (48, 55). The connective function of desmoglein 1 is required for the epidermis to maintain its integrity and structure (57). In addition, the surface proteins SdrC and SasG promote adhesion to squamous epithelium cells, but their binding partners are still unknown (45, 58). Interfering with glycopolymer-receptor interaction might become a new strategy for controlling S. Furthermore, ClfB could represent an ideal target molecule for new decolonization strategies (53). In this context, ClfB can be considered a promising component for the development of a vaccine that would also reduce nasal colonization by S. Direct inhibition can be achieved, for instance, through production of antimicrobials, whereas indirect inhibition may occur via competition for nutrients or modification of living conditions (61). Many nasal Staphylococcus isolates produce antimicrobial substances against bacterial competitors at an unexpectedly high frequency (84%), with S. Importantly, production of many of the antibacterial activities is strongly enhanced or exclusively detectable under specific environmental stress conditions which are present in the human nose, such as hydrogen peroxide release and iron limitation (62). Antimicrobial substances, also called bacteriocins, are categorized into various groups and subgroups (63). Many Staphylococcus isolates are producers of lantibiotics, ribosomally synthesized antimicrobial peptides characterized by the presence of the thioether amino acids lanthionine and methyllanthionine. The anterior and posterior parts of the human nose are lined by different types of epithelial cell, which require alternative bacterial adhesion mechanisms. For the keratinized stratified squamous epithelium in the anterior nasal cavity, S. The corneocytes (desquamated epithelial cells) in the anterior nasal cavity contain high levels of the proteins loricrin, cytokeratin 10, and involucrin (55). Lantibiotics are usually exclusively active against Gram-positive bacteria but often show no activity against S. No compound has been identified that could explain this inhibitory activity; however, the genomes of various P. A recent study showed that secretion of the extracellular serine protease Esp by S. A bacterial strain replacement study highlighted that nasal inoculation of persistent S. The resulting inflammatory response in the nasal epithelium provokes the production of antimicrobial peptides. It is likely that in addition to the processes described above, other factors are involved. It will be important to further investigate which other antimicrobial substances are produced by nasal commensals and which other strategies are used in their competition with S. New findings on nasal colonization may clarify why 20% of the human population is permanently colonized by S. Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation inthe human nose. A poke into the diversity and associations within human anterior nare microbial communities. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Typing of Staphylococcus aureus colonising human nasal carriers by pulsed-field gel electrophoresis. Typing of Staphylococcus epidermidis colonizing in human nares by pulsed-field gel electrophoresis. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Carriage patterns of Staphylococcus aureus in a healthy non-hospital population of adults and children. Staphylococcus aureus nasal carriage is associated with serum 25-hydroxyvitamin D levels, gender and smoking status. Elimination of coincident Staphylococcus aureus nasal and hand carriage with intranasal application of mupirocin calcium ointment. Comparative study of mupirocin and oral co-trimoxazole plus topical fusidic acid in eradication of nasal carriage of methicillin-resistant Staphylococcus aureus. High prevalence of mupirocin resistance in Staphylococcus aureus isolates from a pediatric population. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. An algorithm based on one or two nasal samples is accurate to identify persistent nasal carriers of Staphylococcus aureus. Localization of Staphylococcus aureus in tissue from the nasal vestibule in healthy carriers. Nasal mucus proteomic changes reflect altered immune responses and epithelial permeability in patients with allergic rhinitis. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Immunization with Staphylococcus aureus clumping factor B, a major determinant in nasal carriage, reduces nasal colonization in a murine model. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Two-component anti-Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55.
Generic aricept 10 mg fast delivery
Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting symptoms tuberculosis order aricept with a mastercard. Staphylococcus aureus temperate bacteriophage: carriage and horizontal gene transfer is lineage associated. Sassi M, Augagneur Y, Mauro T, Ivain L, Chabelskaya S, Hallier M, Sallou O, Felden B. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: a longitudinal cohort study based on whole-genome sequencing. Evolution in fast forward: a potential role for mutators in accelerating Staphylococcus aureus pathoadaptation. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y, Katayama Y, Matsuo M, Kuwahara-Arai K, Hishinuma T, Baba T. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Dissemination of novel antimicrobial resistance mechanisms through the insertion sequence mediated spread of metabolic genes. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: temporal changes correlated with frequent loss and gain events. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Risk factors for persistent carriage of methicillin-resistant Staphylococcus aureus. Effect of antibiotics on the bacterial load of meticillin-resistant Staphylococcus aureus colonisation in anterior nares. Impact of antibiotic treatment on the burden of nasal Staphylococcus aureus among hospitalized patients. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. Persistence of livestock-associated methicillinresistant Staphylococcus aureus in field workers after shortterm occupational exposure to pigs and veal calves. Genome plasticity of agrdefective Staphylococcus aureus during clinical infection. Genomic evolution of Staphylococcus aureus isolates colonizing the nares and progressing to bacteremia. Whole-genome sequencing of bloodstream Staphylococcus aureus isolates does not distinguish bacteraemia from endocarditis. Evolutionary trade-offs underlie the multi-faceted virulence of Staphylococcus aureus. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Rapid emergence and evolution of Staphylococcus aureus clones harboring fusC-containing staphylococcal cassette chromosome elements. Epidemiological survey of the first case of vancomycin-resistant Staphylococcus aureus infection in Europe. Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: a three year study in Tehran, Iran. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). Identification of a staphylococcal complement inhibitor with broad host specificity in equid Staphylococcus aureus strains. Development and characterization of a Staphylococcus aureus nasal colonization model in mice. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. Lantibioticmediated anti-lactobacillus activity of a vaginal Staphylococcus aureus isolate. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. It is clear that the acquisition, maintenance, and dissemination of accessory functions have been central to the ongoing success of staphylococci as pathogens. A novel but poorly understood mechanism of staphylococcal genetic exchange, termed mixed-culture transfer or phagemediated conjugation, was described in the 1980s (6, 17, 18). Although phage, or perhaps components of phage, play a role, the process has been shown to be mechanistically distinct from transduction. It is possible that mixed-culture transfer and autotransduction are actually different perspectives on the same phenomenon. Notably, genetic exchange has also been observed to occur more efficiently in vivo than in vitro (23). Such observations suggest that, directly and/or indirectly, the gene transfer mechanisms operating in staphylococci facilitate not only intraspecific transfer, but also interspecific and intergeneric exchange and hence access to an extended and shared reservoir of determinants. Transformation is thought to be limited by extracellular nucleases and is usually very inefficient (6, 7). However, homologs of competence genes from other Gram-positive species are evident in the S. Staphylococci usually possess one or more prophages integrated within their genomes, and generalized transduction is likely to be a frequent mechanism of genetic exchange between strains (9, 10). Staphylococcal conjugative plasmids not only mediate their own self-transfer, but also facilitate the exchange of other coresident nonconjugative plasmids, either by mobilization if the other plasmid carries a relaxation system (11) or more generally via cointegrate formation, and potentially subsequent resolution, in a process termed "conduction" (12, 13). Historically, staphylococcal plasmids can be categorized into one of three main classes based on physical/genetic organization and functional characteristics (24, 25, 30). For example, there are plasmids closely related to multiresistance and conjugative plasmids that lack any recognized resistance genes. However, the rate of plasmid discovery through genome sequencing has rendered incompatibility testing impractical. Unfortunately, coincident identification of additional replicon types (39, 40) has subsequently resulted in discordant classification of some staphylococcal plasmids. Plasmids belonging to each of these four families encode evolutionarily distinct replication initiator (Rep) proteins that possess the conserved domains Rep trans, Rep 1, Rep 2, and RepL, respectively (35). For example, the ssoA carried by pT181 only functions efficiently in staphylococci, whereas the ssoU carried by pE194 family plasmids allows stable replication in a broader range of Grampositive hosts (48). The pC221 MobA relaxase and MobC relaxase accessory proteins are both required for nicking at oriT (54, 55).
10mg aricept free shipping
However medications 5 songs 5 mg aricept with mastercard, SpA proteins can be released from the cell wall by the cell wall hydrolase, LytM (133). The Ig binding domains confer upon SpA the ability to bind the Fcg portion of Igs to prevent opsonization (134). During intravenous infection, SpA prevents opsonophagocytosis of the bacteria by sequestering Igs and impedes the development of specific anti-S. In contrast, isogenic strains that lack spa or express variants that cannot bind to Ig exhibited reduced kidney abscess formation and elicited specific anti-S. Cytotoxic Enzymes b-toxin (also known as b-hemolysin) the b-toxin encoding gene, hlb, is part of the core S. The prophage carries the immune evasion gene cluster encoding for immune evasion factors, such as the staphylococcal complement inhibitor proteins, chemotaxisinhibitory proteins, and staphylokinase (145). The hlb-converting prophage is prevalent in strains associated with human infections (90%), but it is less frequently found in animal isolates (30%) (146). Later, b-toxin was shown to enhance biofilm formation through catalyzing the formation of nucleoprotein matrix in biofilms; therefore, b-toxin is also a biofilm ligase (155). Intranasal administration of b-toxin induces the shedding of syndecan1, a major heparan sulfate proteoglycan molecule on lung epithelial cells, and causes neutrophil infiltration into the lungs in mice (162). Desmogleins are cadherins that are required for desmosome cell-to-cell adhesion to maintain the integrity of the epidermis. Cleavage of Dsg1 disrupts the cell-to-cell adhesion of the epidermis, resulting in blistering and desquamation of the superficial skin. The previous section discussed the mechanisms of toxin-mediated host immune evasion and their roles in S. They can be broadly categorized into two groups: cofactors that activate host zymogens and enzymes for degradation of tissue components (Table 2). While these cofactors and secreted enzymes (exoenzymes) have different substrates and mechanisms of action, they function to break down bacterial and host molecules for nutrient acquisition, bacterial survival, and dissemination. Cofactors for Host Enzyme Activation Coagulase (Coa), von Willebrand factor binding protein (vWbp), and staphylokinase (Sak) are cofactors produced by S. These three proteins hijack different aspects of the host coagulation system, thereby manipulating the host innate defenses to promote bacterial survival and dissemination. Staphylococcal coagulases: Coa and vWbp the ability to induce coagulation is one of the key criteria used in modern medical microbiology for species classification in the genus Staphylococcus-separating coagulase-positive and coagulase-negative species. A majority of staphylococci are coagulase negative, but a few are coagulase positive species, including S. The causative agents, Coa and vWbp, are highly active in coagulating human and rabbit plasma (183). There are 12 isoforms of coa that have been identified thus far; the majority of the variability is attributed to the high sequence variability (>50%) of the N-terminus coding region among different strains (184, 185). In contrast, vwb, which encodes vWbp, is relatively conserved, with only two known alleles (184). The C-terminus of Coa has a 188-residue linker region followed by a repeat region composed of tandem repeats of 27 residues responsible for fibrinogen binding (188, 189). In contrast, the C-terminus of vWbp has a von Willebrand factor binding domain and a fibrinogen binding domain (188, 190). Insertion of the N-terminus of Coa into the Ile16 pocket of prothrombin causes a conformational shift resulting in the activation of the zymogen (189, 191). However, two recently discovered small molecules, argatroband and dabigatran, can inhibit the activity of staphylothrombin (194, 195). The DcoaDvwb strain is less virulent than its wild-type parent, demonstrating a role of the coagulases during infection (187, 196, 197). However, coagulases must be present concurrently with the infecting strain to promote virulence. Ekstedt and Yotis demonstrated that while intracerebral coinjection of purified coagulase with coagulase-negative S. Additionally, Coa is suggested to have a role in the formation of device-associated biofilm formation (199). Coa is required for the formation of pseudocapsule immediately surrounding the abscess, and both vWbp and Coa are required for fibrin formation around the pseudocapsule (187, 200). Sak is produced by lysogenic strains of staphylococci; the prophage encoding Sak typically carries other genes that encode virulence factors such as enterotoxin A and chemotaxis inhibitory proteins (201, 202). The phage carrying sak has also been reported to disrupt the coding sequences of N-acetylmuramyl-L-alanine amidase and peptidoglycan hydrolase (207, 208). Sak can also bind to plasminogen, but this complex is inactive and must be converted to Sak-plasmin to have enzymatic activity (212). In an active Sak-plasmin complex, the first 10 residues at the N-terminus of mature Sak are removed to expose the charged residue, Lys11 (213). Coa and vWbp bind to prothrombin and alter the conformation of the protein to form the complex, staphylothrombin. This complex is highly active and cleaves fibrinogens to fibrins, promoting the formation of fibrinous clots. While Sak binding to plasminogen does not have enzymatic activity, the trimeric complex captures how Sak may bind to plasmin to cleave plasminogen. Sak (orange) is in complex with plasminogen (blue), exposing the catalytic site (red). Sak facilitates the docking of the substrate plasminogen (pink) to promote cleavage by plasmin. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes 651 inactivates Sak (214). The binding of Sak to plasmin directs the active site of plasmin to favor cleavage of the activation loop in plasminogen and promotes the conversion of plasminogen to plasmin by enhancing substrate presentation to plasmin. Circulating Sak-plasmin complexes are sensitive to dissociation by a2-antiplasmin, but fibrin-bound complexes are protected from inactivation (216). The fibrin-bound complexes cleave IgG and human C3b, thus preventing opsonization of the bacteria by the complement system (217). Additionally, Sak-plasmin complexes can activate the matrix metalloprotease 1, which is important for leukocyte migration and activation (218). It is active for human, dog, goat, rabbit, and sheep plasminogen but is inactive for mouse, pig, cow, and buffalo plasminogen (221). Using transgenic mice that produce human plasminogen, studies demonstrated that Sak facilitates S. However, plasmin activation is known to promote wound healing and to reduce inflammation. Thus, during skin infection, Sak may function as a vanguard to establish the primary infection, but after the infection is established, Sak limits the severity of infections to promote dissemination (223). Furthermore, Sak reduces biofilm formation and facilitates the detachment of mature biofilm by activating plasminogen (221). Corroborating these observations, high Sakproducing strains are often associated with less biofilm formation in vitro and noninvasive infections in humans (221, 223). Furthermore, the nuc mutant has decreased fitness during intraperitoneal infection in vivo (232). A mutant expressing only Nuc2 but not Nuc showed that the nuclease is produced during intramuscular infections in mice, albeit at a much lower level (228). Proteases Staphylococci encode three families of secreted proteases: metalloproteases, cysteine proteases, and serine proteases. Collectively, these proteases have roles in nutrient acquisition, bacterial dissemination, and immune evasion. In the following sections, we will discuss the mode of action of each protease family and its proposed role in S. Nuclease requires Ca2+ ions for activity, but not other divalent cations (224, 225). Staphylococcal nuclease is also known as thermonuclease, named after its resistance to heat inactivation (224, 225).
Aricept 10mg line
Food poisoning typically occurs when chromosomal cpe isolates are ingested with food adhd medications 6 year old order generic aricept, while nonfoodborne disease is primarily C. Spo0A likely directly regulates toxin production, because Spo0A directly binds the botE promoter in vitro, which contains a Spo0A box (30). Spo0A represses toxin expression in epidemic 027 ribotypes (6, 76), does not impact toxin expression in the emerging 078 ribotype (76), and variably impacts toxin expression in the historic 012 ribotype (6, 31, 32, 76, 100). However, as with sporulation, nutrient availability strongly influences toxin gene expression since amino acids and glucose repress toxin gene expression through the global regulators CodY and CcpA, respectively (57, 59, 102). RstA inhibits transcription of tcdA and tcdB by directly binding to the promoters and inhibiting the transcription of tcdR and sigD (29; A. Thus, RstA regulates sporulation and toxin gene expression through independent mechanisms (51). However, it remains unclear if this bifunctional protein links sporulation and toxin regulation in the same cell: single-cell analyses would reveal whether toxin-producing cells also sporulate or whether these important processes are asynchronous. Interestingly, the regulatory pathways between sporulation and toxin gene expression may be reciprocal in some C. These sigma factors are essential for sporulation (25, 85, 107) because they coordinate the activation of distinct transcriptional programs within the mother cell and forespore, respectively, that culminate in the formation of a metabolically dormant spore. While the regulation of sporulation-specific sigma factors has been extensively analyzed in B. We first describe the activation and functions of sporulation-specific sigma factors in B. Intercompartmental signaling regulates sporulation sigma factor activation and couples it to specific morphological changes (108). Spo0A induces the transcription of sigF and sigE in the predivisional cell such that sF and sE are present in both the mother cell and forespore, although both sigma factors remain inactive until asymmetric division is complete. This channel, also known as the "feeding tube," is required to maintain transcriptional potential in the forespore (126) and, thus, sG activity. Thus, discrete anti-sigma factors control the activation of sF and sG, respectively, in B. Engulfment increases sG activity, which couples its transcriptional program to morphological changes (131). Thus, proteolytic signaling cascades induced by the forespore activate sE and sK in B. Thus, multiple levels of sK regulation control the precise timing of its activation in B. Transcription factors and sigma factors are shown in bold, and proteins enclosed in boxes directly participate in signaling between the mother cell and forespore (dashed boxes indicate that trans-septum signaling has not been tested yet). Text color denotes whether the factor has been detected at both the transcript and protein level (black), at either the transcript or protein level (purple), or has not been tested yet at the transcript or protein level (blue). Black arrows delineate transcriptional control of gene expression, red arrows indicate signaling pathways, dashed lines indicate that the regulatory relationship remains unknown, and thick arrows demarcate notable points of divergence from the pathway defined in B. Sporulation sigma factor activation and function are further controlled by additional feedback and feedforward loops to ensure that the timing of sporulation gene expression is tightly coordinated with morphological changes (136). Notably, while most of the gene products that control sporulation sigma factor activation in B. Many of the regulatory loops that fine-tune the timing of sporulation sigma factor activation and function in B. Furthermore, the timing of sporulation sigma factor activation does not to appear to be as closely coupled to morphological events as in B. These factors were shown to function in a compartment-specific manner similar to that in B. Notably, sG is present but inactive in the absence of sF (137), indicating that C. Notably, sK appears to function at two stages during sporulation in both these organisms, with the first stage regulating sporulation initiation. Indeed, sigK mutants in both organisms do not appear to initiate asymmetric engulfment (65, 74), unlike in B. This latter phase of gene expression occurs from sF- and sE-dependent promoters (148). Consistent with sK being required early during sporulation in both these organisms, C. Furthermore, sigK mutants in both organisms fail to complete asymmetric engulfment (65, 74), whereas sigE mutants are stalled at this stage (65, 148). While transcriptional analyses indicate that sK acts upstream of sF, Western blot analyses have revealed that sK is not detectable in the absence of sF (86), suggesting that the production (or stability) of these two factors is interdependent. These observations raise the possibility that the sporulation sigma factors exhibit differences in their compartmentspecific activation in C. These observations raise the question of whether sK must be proteolytically activated at this early stage. Summary of Sporulation Regulation Clearly, major differences exist in the functions and regulation of conserved sporulation sigma factors in C. Such studies could provide insight into the evolution of diverse sporulation networks. This section focuses on factors required for engulfment, coat, and exosporium assembly in C. However, in both organisms the channel is essential for maintaining forespore health because the forespore becomes deformed on itself in channel mutants (127, 141). Assembly of the outermost exosporium depends on the BclA collagen-like proteins, which likely create hair-like projections on the spore surface (186, 187), CdeC (185), and CdeM (D. The proteins that make up the outer and inner coat layers are unknown, but CotA and the mucinase, CotE, have been shown to be surface accessible (180, 182). Nevertheless, enzymatic activities have also been determined for several coat proteins (180, 181). For example, alanine racemase interconverts L- and D-alanine (as well as L- and D-serine) and alters the sensitivity of C. Loss of CotE or its C-terminal mucinase domain reduces virulence in a hamster model of infection, indicating that the spore surface actively regulates C. Traces of coat proteins such as CotA, CotB, CotD, and CotE were also observed, which may indicate that they are part of the coat/exosporium interface (178). Sporulation and Germination in Clostridial Pathogens 913 While it is unclear how the C. The cysteine-rich proteins, CdeC and CdeM, have been implicated in exosporium morphogenesis (D. Spores deficient in CdeC have a defective coat that is permeable to lysozyme, have a higher core water content, and are more susceptible to ethanol and heat than wild-type spores (185). Interestingly, cysteine-rich proteins are essential for the morphogenesis of the outer crust of B. Both CdeC and CdeM form dimers, trimers, and higher molecular weight complexes (178, 185, 192), suggesting that a similar self-assembly mechanism might govern the assembly of the outer layers of C. Although many germination-related proteins are conserved in the clostridial pathogens, notable differences in their function and mechanisms of action have been identified in C. As discussed below, the order in which cortex hydrolysis and core hydration occurs differs between these species and is even strainspecific in the case of C. Germinant Sensing and Signaling Environmental signals In most spore-forming bacteria, germinants are nutrient signals such as amino acids, monosacharides, nucleosides, salts, and organic acids (28). Taurocholate is the most potent of the cholate-derived germinants, while chenodeoxycholate is an efficient competitive inhibitor of taurocholate-mediated germination (201). L-alanine, L-cysteine, L-methionine, L-serine, L-phenylalanine, and glycine can induce spore germination of group I proteolytic C. Germinant selectivity is likely influenced by adaptation to specific environmental niches. While classical germinants are directly sensed through germinant receptors, nonnutrient germinants can artificially trigger germination of many bacterial species independent of Exosporium Function in C. Notably, two exosporium morphotypes are observed in clonal populations: thin and thick electrondense layers, although both have hair-like extensions (197, 198).